
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,293,154 – 6,293,251 |
| Length | 97 |
| Max. P | 0.947925 |

| Location | 6,293,154 – 6,293,251 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.78 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -28.27 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
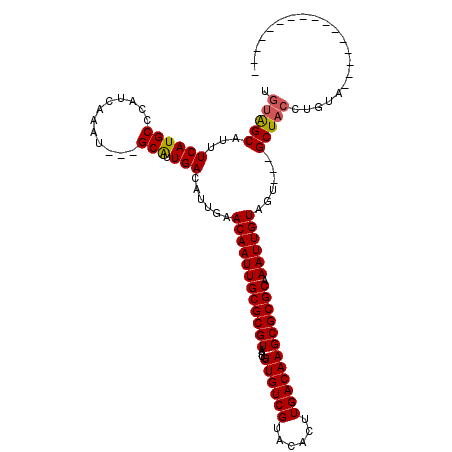
>3L_DroMel_CAF1 6293154 97 + 23771897 AAAGGCAUUUCAUGCUCAUCAAAU---GCGUUGACAUUGAACAAUUGCGCGUAAUGUUGUCGUACACUUGACAAGCGCGCAAAUUGUAGU---GCUACCUGUA---------------- ...((((((((((((.........---))).)))......((((((((((((....((((((......)))))))))))).)))))))))---))).......---------------- ( -28.20) >DroVir_CAF1 72381 97 + 1 UGUAGCAUUUCAUGCCCAUCAAAU---GCAUUGACAUUGAACAAUUGCGCGUAAUGUUGUCGUACACUUGACAAGCGCGCAAAUUGUAGU---GCUACCUGUG---------------- .((((((((((((((.........---))).)))......((((((((((((....((((((......)))))))))))).)))))))))---))))).....---------------- ( -31.20) >DroPse_CAF1 86373 119 + 1 AGUGGCAUUUCAUGCUCAUCAAAUGCUGCGUUGACAUUGAACAAUUGCGCGUAAUGUUGUCGUACACUUGACAAGCGCGCAAAUUGUAGUGCUGCUACCUGUAGCUGUAGCUGUAGCUG .(((((((...)))).))).....(((((((((.((....((((((((((((....((((((......)))))))))))).))))))...(((((.....))))))))))).))))).. ( -39.40) >DroGri_CAF1 63049 97 + 1 UGUAGCAUUUCAUGCCCAUCAAAU---GCAUUGACAUUGAACAAUUGCGCGUAAUGUUGUCGUACACUUGACAAGCGCGCAAAUUGUAGU---GCUACCUGUG---------------- .((((((((((((((.........---))).)))......((((((((((((....((((((......)))))))))))).)))))))))---))))).....---------------- ( -31.20) >DroMoj_CAF1 69956 97 + 1 UGUAGCAUUUCAUGCCCAUCAAAU---GCAUUGACAUUGAACAAUUGCGCGUAAUGUUGUCGUACACUUGACAAGCGCGCAAAUUGUAGU---GCUACCUGUG---------------- .((((((((((((((.........---))).)))......((((((((((((....((((((......)))))))))))).)))))))))---))))).....---------------- ( -31.20) >DroPer_CAF1 89805 119 + 1 UGUGGCAUUUCAUGCUCAUCAAAUGCUGCGUUGACAUUGAACAAUUGCGCGUAAUGUUGUCGUACACUUGACAAGCGCGCAAAUUGUAGAGCUGCUACCUGUAGCUGUAGCUGUAGCUG .(((((((...)))).))).....(((((((((.......((((((((((((....((((((......)))))))))))).))))))..((((((.....)))))).)))).))))).. ( -40.30) >consensus UGUAGCAUUUCAUGCCCAUCAAAU___GCAUUGACAUUGAACAAUUGCGCGUAAUGUUGUCGUACACUUGACAAGCGCGCAAAUUGUAGU___GCUACCUGUA________________ .(((((...((((((............))).)))......((((((((((((....((((((......)))))))))))).))))))......)))))..................... (-28.27 = -28.10 + -0.17)

| Location | 6,293,154 – 6,293,251 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.78 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -23.27 |
| Energy contribution | -22.77 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
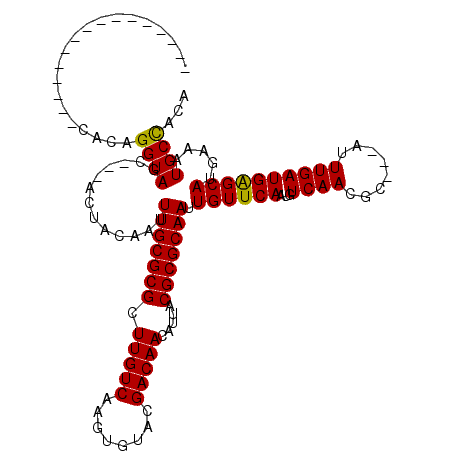
>3L_DroMel_CAF1 6293154 97 - 23771897 ----------------UACAGGUAGC---ACUACAAUUUGCGCGCUUGUCAAGUGUACGACAACAUUACGCGCAAUUGUUCAAUGUCAACGC---AUUUGAUGAGCAUGAAAUGCCUUU ----------------...(((((((---.(........).))(((..((((((((..((((.....(((......)))....))))...))---))))))..)))......))))).. ( -27.40) >DroVir_CAF1 72381 97 - 1 ----------------CACAGGUAGC---ACUACAAUUUGCGCGCUUGUCAAGUGUACGACAACAUUACGCGCAAUUGUUCAAUGUCAAUGC---AUUUGAUGGGCAUGAAAUGCUACA ----------------.....(((((---(...(((((.(((((.(((((........))))).....))))))))))((((.((((....(---....)...)))))))).)))))). ( -28.30) >DroPse_CAF1 86373 119 - 1 CAGCUACAGCUACAGCUACAGGUAGCAGCACUACAAUUUGCGCGCUUGUCAAGUGUACGACAACAUUACGCGCAAUUGUUCAAUGUCAACGCAGCAUUUGAUGAGCAUGAAAUGCCACU .((((........))))...((((((.(((........)))))(((..((((((((.((...(((((..(((....)))..)))))...))..))))))))..)))......))))... ( -32.30) >DroGri_CAF1 63049 97 - 1 ----------------CACAGGUAGC---ACUACAAUUUGCGCGCUUGUCAAGUGUACGACAACAUUACGCGCAAUUGUUCAAUGUCAAUGC---AUUUGAUGGGCAUGAAAUGCUACA ----------------.....(((((---(...(((((.(((((.(((((........))))).....))))))))))((((.((((....(---....)...)))))))).)))))). ( -28.30) >DroMoj_CAF1 69956 97 - 1 ----------------CACAGGUAGC---ACUACAAUUUGCGCGCUUGUCAAGUGUACGACAACAUUACGCGCAAUUGUUCAAUGUCAAUGC---AUUUGAUGGGCAUGAAAUGCUACA ----------------.....(((((---(...(((((.(((((.(((((........))))).....))))))))))((((.((((....(---....)...)))))))).)))))). ( -28.30) >DroPer_CAF1 89805 119 - 1 CAGCUACAGCUACAGCUACAGGUAGCAGCUCUACAAUUUGCGCGCUUGUCAAGUGUACGACAACAUUACGCGCAAUUGUUCAAUGUCAACGCAGCAUUUGAUGAGCAUGAAAUGCCACA .((((........))))...((((.((((((.((((((.(((((.(((((........))))).....)))))))))))(((((((.......))).)))).)))).))...))))... ( -32.80) >consensus ________________CACAGGUAGC___ACUACAAUUUGCGCGCUUGUCAAGUGUACGACAACAUUACGCGCAAUUGUUCAAUGUCAACGC___AUUUGAUGAGCAUGAAAUGCCACA ....................((((.............(((((((.(((((........))))).....))))))).((((((...((((........)))))))))).....))))... (-23.27 = -22.77 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:04 2006