
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,258,688 – 6,258,860 |
| Length | 172 |
| Max. P | 0.997311 |

| Location | 6,258,688 – 6,258,781 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -13.00 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6258688 93 + 23771897 ACACAAAAAAAA--AGUAAAAAAUGUCGGGAGGUGUGGCCAAUUGUAAAGGCUUGCAAAGC--------AUUUGACACAUUGGAAACAUGUGGUAACAUGCAA ............--.((((...((..(....)..)).(((.........)))))))...((--------(((((.(((((.(....)))))).))).)))).. ( -19.30) >DroSec_CAF1 25406 81 + 1 ACAAA--------------AAAAUGUCGGGAGGUGUGGCCAAUUGUAAAGGCUGGCAAAGC--------AUUUGACACAUUGGCAACAUGUGGCAACAUGCAA .....--------------....(((......((((.(((((((((....(((.....)))--------.....))).)))))).))))(((....)))))). ( -20.90) >DroSim_CAF1 26005 73 + 1 ACAAAAA------------AAAAUGUCGGGAGGUGUGGCCAAUUGUAAAGGCUUGCAAAGC--------AUUUGACACAUUGGCA----------ACAUGCAA .......------------.............((((.(((((((((....(((.....)))--------.....))).)))))).----------)))).... ( -16.20) >DroEre_CAF1 25751 99 + 1 ACAAAAAGAAAC----AGAAAAAUGUCGGGAGGUGUGGUCAUUUGUAAAGGCUGGCAAAGCGCAGCCACAUUUGACACAUUGGCAACAUGUGGCAACAUGCAA ............----.......(((((....(((((.(((..(((...(((((((...)).))))))))..))))))))))))).(((((....)))))... ( -26.50) >DroYak_CAF1 29149 95 + 1 ACAAAAAGAAAACGAGAAAAAAAUGUCAGGAGGUGUGGUCAAUUGUAGAGGCUGGCAAAGC--------AUUUGACACAUUGGCAACAUGUGGCAACAUGCAA .......................((((((....((..(....)..))....))))))..((--------(((((.(((((.(....)))))).))).)))).. ( -19.40) >consensus ACAAAAA_AAA_______AAAAAUGUCGGGAGGUGUGGCCAAUUGUAAAGGCUGGCAAAGC________AUUUGACACAUUGGCAACAUGUGGCAACAUGCAA .......................((((((...((((((((.........))))..((((...........)))))))).)))))).(((((....)))))... (-13.00 = -13.80 + 0.80)

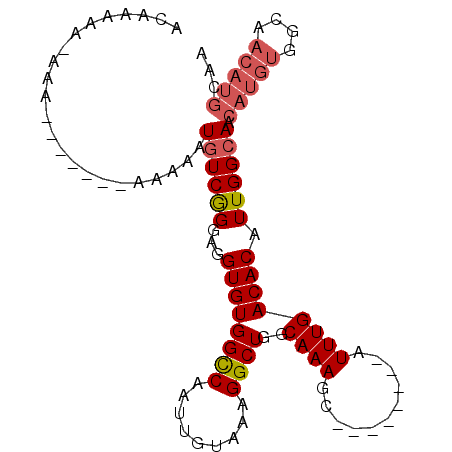
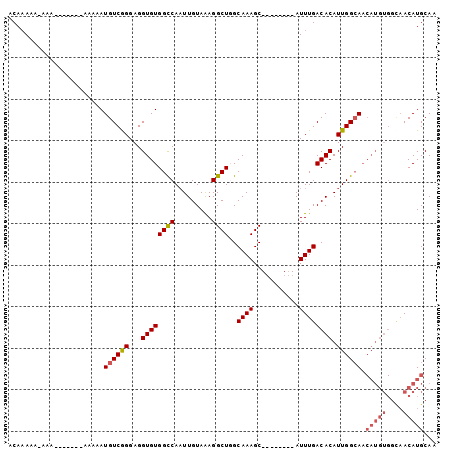
| Location | 6,258,722 – 6,258,821 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.85 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -8.91 |
| Energy contribution | -9.49 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6258722 99 + 23771897 GGCCAAUUGUAAAGGCUUGCAAAGC--------AUUUGACACAUUGGAAACAUGU----GGUAACAUGCAAAUGUUGCAACGAUACCAUACCACAUUACGAUUGCUAAAUU (((.((((((((.((((.....)))--------..(((.(((((.(....)))))----)(((((((....)))))))..)))..........).)))))))))))..... ( -25.80) >DroSec_CAF1 25428 94 + 1 GGCCAAUUGUAAAGGCUGGCAAAGC--------AUUUGACACAUUGGCAACAUGU----GGCAACAUGCAAAUGUUGCAA-----CCAUACCACAUUACGAUUGCUAAAUU (((.((((((((.((((.....)))--------......(((((.(....)))))----)(((((((....)))))))..-----........).)))))))))))..... ( -28.00) >DroSim_CAF1 26029 84 + 1 GGCCAAUUGUAAAGGCUUGCAAAGC--------AUUUGACACAUUGGCA--------------ACAUGCAAAUGUUGCAA-----CCAUACCACAUUACGAUUGCUAAAUU (((.((((((((.((.(((((..((--------(((((.((...((...--------------.))))))))))))))))-----))........)))))))))))..... ( -21.90) >DroEre_CAF1 25783 101 + 1 GGUCAUUUGUAAAGGCUGGCAAAGCGCAGCCACAUUUGACACAUUGGCAACAUGU----GGCAACAUGCAAAUGUUGCAA-----CCAUGCCACAUUACGUUUGCUAAAU- .((((..(((...(((((((...)).))))))))..))))...((((((((((((----((((...((((.....)))).-----...))))))))...).)))))))..- ( -36.70) >DroYak_CAF1 29185 94 + 1 GGUCAAUUGUAGAGGCUGGCAAAGC--------AUUUGACACAUUGGCAACAUGU----GGCAACAUGCAAAUGUUGCAA-----CCUUACCACAUUACGUUGGAUAAAUA .(((...(((((.((((.....)))--------......(((((.(....)))))----)(((((((....)))))))..-----........).)))))...)))..... ( -23.90) >DroAna_CAF1 25064 82 + 1 -----------AACGCCGGCAACAC--------AUUUAACAAAUUGGCAACAUGUCGCUGGCAACAUGCAAAUGUUGCAA-----CGGGUUCAGAUCG-----AGUAAAUG -----------......(....).(--------(((((........(((((((...((((....)).))..)))))))..-----(((.......)))-----..)))))) ( -19.20) >consensus GGCCAAUUGUAAAGGCUGGCAAAGC________AUUUGACACAUUGGCAACAUGU____GGCAACAUGCAAAUGUUGCAA_____CCAUACCACAUUACGAUUGCUAAAUU .......(((((.((((.....)))...................(((.............(((((((....)))))))............)))).)))))........... ( -8.91 = -9.49 + 0.59)



| Location | 6,258,747 – 6,258,860 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.39 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -7.96 |
| Energy contribution | -8.21 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6258747 113 + 23771897 AUUUGACACAUUGGAAACAUGU----GGUAACAUGCAAAUGUUGCAACGAUACCAUACCACAUUACGAUUGCUAAAUUAAAAAUGUGGUACA-UUAUGGUACAAUGCCAAUGACUACG ........((((((.....(((----.(((((((....))))))).))).((((((((((((((..((((....))))...)))))))))..-...))))).....))))))...... ( -31.30) >DroSec_CAF1 25453 108 + 1 AUUUGACACAUUGGCAACAUGU----GGCAACAUGCAAAUGUUGCAA-----CCAUACCACAUUACGAUUGCUAAAUUAAAAAUGUGGUACA-UUAUGGUACAAUGCCAAUGACUGCG ........((((((((...(((----.(((((((....))))))).(-----((((((((((((..((((....))))...)))))))))..-...))))))).))))))))...... ( -35.60) >DroSim_CAF1 26054 98 + 1 AUUUGACACAUUGGCA--------------ACAUGCAAAUGUUGCAA-----CCAUACCACAUUACGAUUGCUAAAUUAAAAAUGUGGUACA-UUAUGGUACAAUGCCAAUGAGUACG .....((.((((((((--------------...((((.....))))(-----((((((((((((..((((....))))...)))))))))..-...))))....)))))))).))... ( -30.00) >DroEre_CAF1 25816 88 + 1 AUUUGACACAUUGGCAACAUGU----GGCAACAUGCAAAUGUUGCAA-----CCAUGCCACAUUACGUUUGCUAAAU-GACUACGAA-CAUAGAUAUUU------------------- ........((((((((((((((----((((...((((.....)))).-----...))))))))...).))))).)))-)........-...........------------------- ( -20.60) >DroYak_CAF1 29210 100 + 1 AUUUGACACAUUGGCAACAUGU----GGCAACAUGCAAAUGUUGCAA-----CCUUACCACAUUACGUUGGAUAAAUAGAAUAUGAA-AAUA-AUACUUAACAAUACCUA-------G (((..((((((.(....)))))----.(((((((....)))))))..-----..............))..)))........(((...-.)))-.................-------. ( -18.20) >DroAna_CAF1 25078 93 + 1 AUUUAACAAAUUGGCAACAUGUCGCUGGCAACAUGCAAAUGUUGCAA-----CGGGUUCAGAUCG-----AGUAAAUGGAAUAUUU-------AUUCUUU-UAAUAUCUU-------G .............(((((((...((((....)).))..)))))))..-----.((((...((..(-----((((((((...)))))-------))))..)-)...)))).-------. ( -20.60) >consensus AUUUGACACAUUGGCAACAUGU____GGCAACAUGCAAAUGUUGCAA_____CCAUACCACAUUACGAUUGCUAAAUUAAAAAUGUG_UACA_AUAUGGUACAAUGCCAA_______G ...........(((.............(((((((....)))))))............))).......................................................... ( -7.96 = -8.21 + 0.25)



| Location | 6,258,747 – 6,258,860 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.39 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -6.85 |
| Energy contribution | -8.10 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6258747 113 - 23771897 CGUAGUCAUUGGCAUUGUACCAUAA-UGUACCACAUUUUUAAUUUAGCAAUCGUAAUGUGGUAUGGUAUCGUUGCAACAUUUGCAUGUUACC----ACAUGUUUCCAAUGUGUCAAAU ........(((((((.........(-(((((((((((.................)))))))))(((((.(((.(((.....)))))).))))----)))).........))))))).. ( -26.80) >DroSec_CAF1 25453 108 - 1 CGCAGUCAUUGGCAUUGUACCAUAA-UGUACCACAUUUUUAAUUUAGCAAUCGUAAUGUGGUAUGG-----UUGCAACAUUUGCAUGUUGCC----ACAUGUUGCCAAUGUGUCAAAU .(((..((((((((.(((((((...-..(((((((((.................))))))))))))-----).(((((((....))))))).----)))...)))))))))))..... ( -32.13) >DroSim_CAF1 26054 98 - 1 CGUACUCAUUGGCAUUGUACCAUAA-UGUACCACAUUUUUAAUUUAGCAAUCGUAAUGUGGUAUGG-----UUGCAACAUUUGCAUGU--------------UGCCAAUGUGUCAAAU .(.((.((((((((....((((...-..(((((((((.................))))))))))))-----)((((.....))))...--------------)))))))).))).... ( -27.73) >DroEre_CAF1 25816 88 - 1 -------------------AAAUAUCUAUG-UUCGUAGUC-AUUUAGCAAACGUAAUGUGGCAUGG-----UUGCAACAUUUGCAUGUUGCC----ACAUGUUGCCAAUGUGUCAAAU -------------------((((..((((.-...))))..-)))).((((.....((((((((...-----.((((.....))))...))))----)))).))))............. ( -22.50) >DroYak_CAF1 29210 100 - 1 C-------UAGGUAUUGUUAAGUAU-UAUU-UUCAUAUUCUAUUUAUCCAACGUAAUGUGGUAAGG-----UUGCAACAUUUGCAUGUUGCC----ACAUGUUGCCAAUGUGUCAAAU .-------..(((((((.((((((.-.((.-.....))..))))))..)))....(((((((((.(-----(.(((.....))))).)))))----))))..))))............ ( -21.60) >DroAna_CAF1 25078 93 - 1 C-------AAGAUAUUA-AAAGAAU-------AAAUAUUCCAUUUACU-----CGAUCUGAACCCG-----UUGCAACAUUUGCAUGUUGCCAGCGACAUGUUGCCAAUUUGUUAAAU .-------.((((..((-((.((((-------....))))..))))..-----..)))).(((..(-----(((((((((((((.((....)))))).)))))).))))..))).... ( -15.30) >consensus C_______UUGGCAUUGUAAAAUAA_UGUA_CACAUAUUCAAUUUAGCAAUCGUAAUGUGGUAUGG_____UUGCAACAUUUGCAUGUUGCC____ACAUGUUGCCAAUGUGUCAAAU ..........................................................(((((..........(((((((....)))))))...........)))))........... ( -6.85 = -8.10 + 1.25)
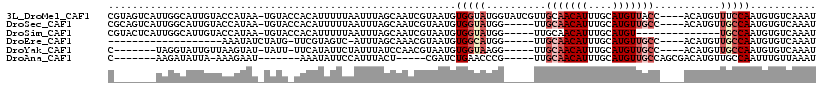


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:58 2006