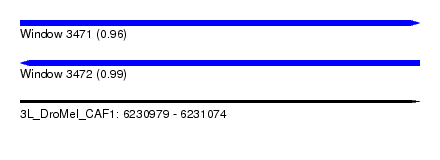
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,230,979 – 6,231,074 |
| Length | 95 |
| Max. P | 0.994041 |

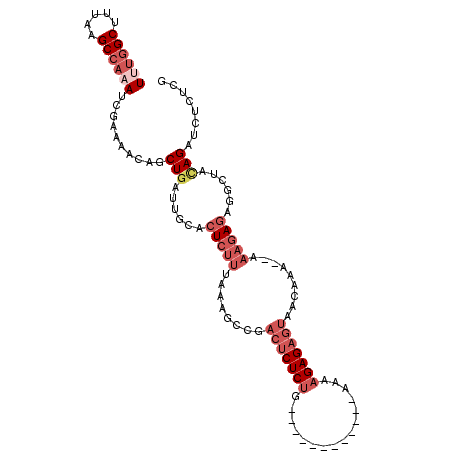
| Location | 6,230,979 – 6,231,074 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6230979 95 + 23771897 CUAGAGAUCUAUAUACUCUCUU---UUUGGUACUCUCUUCU------------CAGAGAGUCGGCUUUAAAGAGUGCAAUCAGCUGUUUUUGAUUUGGCUUAAAGCCAAA .(((((((.........(((((---(..((((((((((...------------.)))))))..)))..)))))).((.....)).))))))).((((((.....)))))) ( -25.40) >DroSec_CAF1 21180 97 + 1 CGAGAUAUCUAUAGCCUCUCUUU-UUUUGUUACUCUCUUUU------------CAGAGAGUCGGCUUUAAAGAGCGCAAUCAGCUGUUUUCGAUUUGGCUUAAAGCCAAA (((((.....((((((.((((((-....((((((((((...------------.))))))).)))...)))))).)......)))))))))).((((((.....)))))) ( -27.20) >DroSim_CAF1 21670 98 + 1 CGAGAGAUCUUUAGCCUCUCUUUUUUUUGUUACUCUCUUUU------------CAGAGAGUCGGCUUUAAAGAGCGCAAUCAGCUGUUUUCGAUUUGGCUUAAAGCCAAA ((((((..((...((..((((((.....((((((((((...------------.))))))).)))...)))))).))....))...)))))).((((((.....)))))) ( -26.70) >DroEre_CAF1 22602 95 + 1 CGAAA-AUCUGUAGCCUCUCUUU--UUUGGUACUCUCUUUU------------CAGAGAGUCGGCUUUCAAGAGGGCAAUCAGCUGUUUUCGAUUUGGCUUAAAGCCAGA (((((-(((((..(((.(((((.--...((((((((((...------------.)))))))..)))...))))))))...)))..))))))).((((((.....)))))) ( -34.80) >DroYak_CAF1 21424 96 + 1 UGAAAGAUCUGUAGCUUAUCUUU--UUUGAUACUCUCUUUU------------CAGAGAGUCGGCUUUAAAGAGUGUAAUCAGCUGUUUUCGAUUUGGCUUAAAGCCAAA .(((((..(((..((...(((((--...(..(((((((...------------.)))))))...)...)))))..))...)))...)))))..((((((.....)))))) ( -25.20) >DroAna_CAF1 22899 108 + 1 GGGAGAGUCUGCAGCCUCUCGUU--UUGAACCUGCUCUUUCUACCAGCUGUGACUGAGAGCCGGCUUUGUUCAGUUUAAUCAGCUGAUUCGGUUUUGGCCUUAAGCGCGA .((((((...((((..((.....--..))..)))).))))))......((((..((((.(((((..(((.((((((.....))))))..)))..)))))))))..)))). ( -32.50) >consensus CGAAAGAUCUGUAGCCUCUCUUU__UUUGAUACUCUCUUUU____________CAGAGAGUCGGCUUUAAAGAGUGCAAUCAGCUGUUUUCGAUUUGGCUUAAAGCCAAA (((((..........................(((((((................)))))))(((((...............))))).))))).((((((.....)))))) (-14.37 = -14.57 + 0.20)

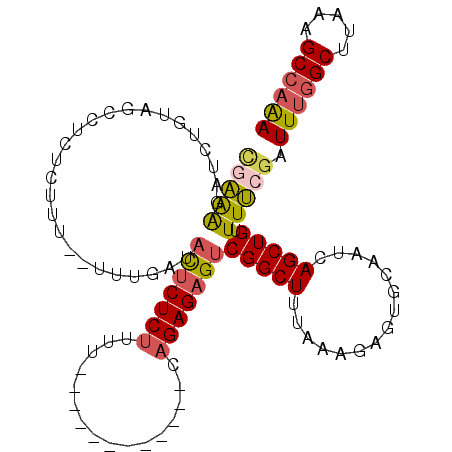
| Location | 6,230,979 – 6,231,074 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -13.87 |
| Energy contribution | -15.87 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6230979 95 - 23771897 UUUGGCUUUAAGCCAAAUCAAAAACAGCUGAUUGCACUCUUUAAAGCCGACUCUCUG------------AGAAGAGAGUACCAAA---AAGAGAGUAUAUAGAUCUCUAG ..((((((((((...(((((........)))))......))))))))))(((((((.------------...)))))))......---.(((((.........))))).. ( -26.40) >DroSec_CAF1 21180 97 - 1 UUUGGCUUUAAGCCAAAUCGAAAACAGCUGAUUGCGCUCUUUAAAGCCGACUCUCUG------------AAAAGAGAGUAACAAAA-AAAGAGAGGCUAUAGAUAUCUCG ..((((((((((((((.(((........)))))).)))...))))))))(((((((.------------...))))))).......-...((((..(....)...)))). ( -28.90) >DroSim_CAF1 21670 98 - 1 UUUGGCUUUAAGCCAAAUCGAAAACAGCUGAUUGCGCUCUUUAAAGCCGACUCUCUG------------AAAAGAGAGUAACAAAAAAAAGAGAGGCUAAAGAUCUCUCG ..((((((((((((((.(((........)))))).)))...))))))))(((((((.------------...)))))))...........(((((((....).)))))). ( -32.00) >DroEre_CAF1 22602 95 - 1 UCUGGCUUUAAGCCAAAUCGAAAACAGCUGAUUGCCCUCUUGAAAGCCGACUCUCUG------------AAAAGAGAGUACCAAA--AAAGAGAGGCUACAGAU-UUUCG ..((((.....))))...((((((...(((...((((((((........(((((((.------------...)))))))......--.))))).)))..))).)-))))) ( -34.86) >DroYak_CAF1 21424 96 - 1 UUUGGCUUUAAGCCAAAUCGAAAACAGCUGAUUACACUCUUUAAAGCCGACUCUCUG------------AAAAGAGAGUAUCAAA--AAAGAUAAGCUACAGAUCUUUCA ((((((.....))))))..((((....(((.......(((((.......(((((((.------------...)))))))......--))))).......)))...)))). ( -24.66) >DroAna_CAF1 22899 108 - 1 UCGCGCUUAAGGCCAAAACCGAAUCAGCUGAUUAAACUGAACAAAGCCGGCUCUCAGUCACAGCUGGUAGAAAGAGCAGGUUCAA--AACGAGAGGCUGCAGACUCUCCC ....((((..((......))..((((((((.....(((((....((....)).)))))..)))))))).....))))........--...(((((........))))).. ( -26.30) >consensus UUUGGCUUUAAGCCAAAUCGAAAACAGCUGAUUGCACUCUUUAAAGCCGACUCUCUG____________AAAAGAGAGUAACAAA__AAAGAGAGGCUACAGAUCUCUCG ((((((.....))))))..........(((......(((((........(((((((................))))))).........)))))......)))........ (-13.87 = -15.87 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:41 2006