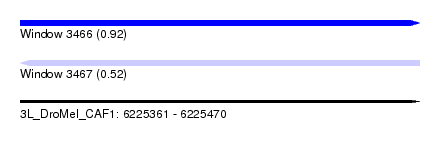
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,225,361 – 6,225,470 |
| Length | 109 |
| Max. P | 0.918255 |

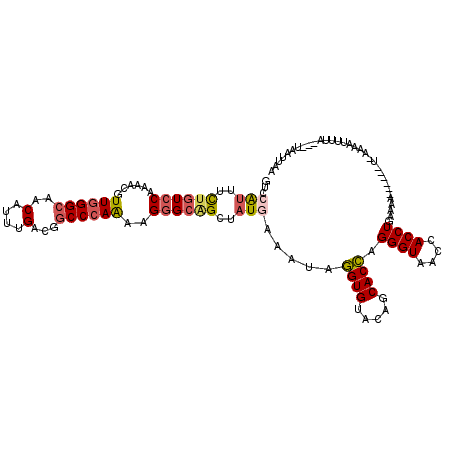
| Location | 6,225,361 – 6,225,470 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -32.71 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6225361 109 + 23771897 GUCCACUUCUGUCCAGAACGUUGGGCAACAUUUGACGGCCCAAAAGGGCAGCUAUGAAAUAGGUGUACAGCACCAGGGUGACUACCUGAAAA------U-AAAGUUUUA---UAAUUAA (((......(((((((....)))))))......))).((((....))))...((((((((.((((.....)))).((((....)))).....------.-...))))))---))..... ( -30.30) >DroGri_CAF1 15742 112 + 1 UGCCAUUACGAUCCAAGAUAUUUGGCAACAUUUGGCGACCCAACAGUGCGGCUAUAAAAUACGUGUAGAGCACGAGGGUAACAACCUACAA------AU-AGUAAUUUGAAUUAAUUAA .((((.......(((((...))))).......)))).((((....((((..(((((.......))))).))))..))))..(((..(((..------..-.)))..))).......... ( -25.04) >DroSec_CAF1 15598 109 + 1 GUCCAUUUCUGUCCAGAACGUUGGGCAACAUUUGACGGCCCAAAAGGGCAGCGAUGAAAUAGGUGUACAGCACCAGGGUAACCACCUGAAAA------U-AAAGUUUUA---UAAUUAA ...((((.((((((......((((((..(....)...))))))..)))))).)))).....((((.....)))).((((....)))).....------.-.........---....... ( -29.30) >DroEre_CAF1 17055 116 + 1 GUCCAUUUCUGUCCAAGACGUUGGGCAACAUUUGACGGCCCAAGAGGGCAGCUAUGAAAUAGGUGUACAGCACCAGGGUGACCACCUGGAAAUGGAAAUGGAAAUUCGA---UGAUUUA .(((((((((((((((....))))))).(((((....((((....))))..(((.(.....((((.....)))).((....)).).)))))))))))))))).......---....... ( -39.40) >DroYak_CAF1 15744 110 + 1 GUCCAUUUCUGUCCAAAACGUUGGGCAACAUUUGGCGGCCCAAAAGGGCAGCUAUGAAAUAGGUGUACAGCACCAGGGUGACCACCUAAAAA------UGGAAAUUUUA---UGAUUUC .(((((((.(((((((....))))))).....((((.((((....)))).)))).......((((.....)))).((....))......)))------)))).......---....... ( -36.60) >DroAna_CAF1 16657 112 + 1 GGCCGUUUUUGUCCAAGACAUUGGGCAGCAUCUGGCGGCCCAGAAGGGCGGCUAUAAAGUAGGUGUACAGCACCAGGGUAACCACCUGAAAA-----AU-AAUAUUUCA-CUUAAUUAA (((((((((((((...))).((((((.((.....)).))))))))))))))))...((((.((((.....)))).((((....)))).....-----..-........)-)))...... ( -35.60) >consensus GUCCAUUUCUGUCCAAAACGUUGGGCAACAUUUGACGGCCCAAAAGGGCAGCUAUGAAAUAGGUGUACAGCACCAGGGUAACCACCUGAAAA______U_AAAAUUUUA___UAAUUAA ...(((..((((((......((((((..(....)...))))))..))))))..))).....((((.....)))).((((....))))................................ (-21.50 = -22.03 + 0.53)

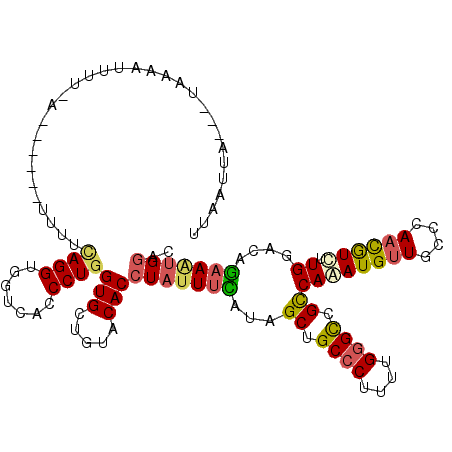
| Location | 6,225,361 – 6,225,470 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6225361 109 - 23771897 UUAAUUA---UAAAACUUU-A------UUUUCAGGUAGUCACCCUGGUGCUGUACACCUAUUUCAUAGCUGCCCUUUUGGGCCGUCAAAUGUUGCCCAACGUUCUGGACAGAAGUGGAC .......---.........-.------........(((.(((....)))))).....(((((((......((((....)))).(((.((((((....))))))...))).))))))).. ( -25.60) >DroGri_CAF1 15742 112 - 1 UUAAUUAAUUCAAAUUACU-AU------UUGUAGGUUGUUACCCUCGUGCUCUACACGUAUUUUAUAGCCGCACUGUUGGGUCGCCAAAUGUUGCCAAAUAUCUUGGAUCGUAAUGGCA .......((((((..(((.-..------..)))((((((......((((.....))))......))))))......)))))).((((.(((...((((.....))))..)))..)))). ( -23.90) >DroSec_CAF1 15598 109 - 1 UUAAUUA---UAAAACUUU-A------UUUUCAGGUGGUUACCCUGGUGCUGUACACCUAUUUCAUCGCUGCCCUUUUGGGCCGUCAAAUGUUGCCCAACGUUCUGGACAGAAAUGGAC .......---.........-.------....((((.(....)))))(((.....)))(((((((......((((....)))).(((.((((((....))))))...))).))))))).. ( -26.80) >DroEre_CAF1 17055 116 - 1 UAAAUCA---UCGAAUUUCCAUUUCCAUUUCCAGGUGGUCACCCUGGUGCUGUACACCUAUUUCAUAGCUGCCCUCUUGGGCCGUCAAAUGUUGCCCAACGUCUUGGACAGAAAUGGAC .......---.......((((((((.....(((((.(....)))))).(((((...........))))).((((....)))).(((..(((((....)))))....))).)))))))). ( -32.20) >DroYak_CAF1 15744 110 - 1 GAAAUCA---UAAAAUUUCCA------UUUUUAGGUGGUCACCCUGGUGCUGUACACCUAUUUCAUAGCUGCCCUUUUGGGCCGCCAAAUGUUGCCCAACGUUUUGGACAGAAAUGGAC .......---.......((((------((((((((((..(((....))).....)))))..((((..((.((((....)))).)).(((((((....))))))))))).))))))))). ( -32.20) >DroAna_CAF1 16657 112 - 1 UUAAUUAAG-UGAAAUAUU-AU-----UUUUCAGGUGGUUACCCUGGUGCUGUACACCUACUUUAUAGCCGCCCUUCUGGGCCGCCAGAUGCUGCCCAAUGUCUUGGACAAAAACGGCC .((((((..-(((((....-..-----.)))))..))))))..(((((((((((.........)))))).((((....)))).)))))..((((.....((((...))))....)))). ( -29.70) >consensus UUAAUUA___UAAAAUUUU_A______UUUUCAGGUGGUCACCCUGGUGCUGUACACCUAUUUCAUAGCUGCCCUUUUGGGCCGCCAAAUGUUGCCCAACGUCUUGGACAGAAAUGGAC ...............................((((.......))))(((.....)))(((((((...((.((((....)))).))((((((((....)))))).))....))))))).. (-18.34 = -18.35 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:37 2006