
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,221,347 – 6,221,458 |
| Length | 111 |
| Max. P | 0.792162 |

| Location | 6,221,347 – 6,221,458 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.92 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.03 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6221347 111 - 23771897 --------CUACUGCACCAGGAGAGCGGUCAGGUGCUGGGCACGCUGCUGCUCUCGC-CCAUCAAAGAGAUGGAUAGCUCAUCGUCCAAUAACACUCUGAUUCCAGGUCAUCCGGCUACC --------...(((...(((((((((((.(((((((...)))).)))))))))))((-(((((.....)))))...))..................)))....)))(((....))).... ( -34.80) >DroVir_CAF1 11832 111 - 1 --------AAUUUGCACCAGGACAGCGGCCAGGUGCUCGGCACACUGCUGCUGUCAC-CCAUCAAAGAAAUGGAUAGCUCAUCAUCCAAUAAUACUCUGAUUCCAGGGCAUCCAGCCACA --------.....((.....(((((((((.((((((...)))).)))))))))))..-............(((((.((((((((.............))))....))))))))))).... ( -34.02) >DroPse_CAF1 15645 120 - 1 GCAUGCAUCUGGGGCACGAUGGCGGCGGCCAGGUGCUGGGCACGCUCCUCCUGUCGCCCCAUCAAAGAGAUGGACAGCUCCUCAUCGAAUAACACUCUGAUUCCAGGACACCCAGCCACA ........(((((...(((((((((((((.(((....(((....)))..)))))))))(((((.....)))))...))....)))))........((((....))))...)))))..... ( -39.40) >DroGri_CAF1 11755 111 - 1 --------AAUUUGCACCAGGACAGCGGCCAGGUGCUGGGCACACUGCUGCUGUCGC-CCAUCAAAGAGAUGGAUAGCACAUCAACCAAUAACACUCUGAUUCCAGGGCACCCAGCAACA --------...((((..((((((((((((.((((((...)))).)))))))))))((-(((((.....)))))...))..................)))......((....)).)))).. ( -38.50) >DroWil_CAF1 12660 112 - 1 ------AGCUGGAGCAC-AGGAUAGUGGCCAAGUGCUAGGCACCUUGCUAUUGUCAC-CCAUUAAAGAGAUGGAUAGUUCAUCAUCGAAUAACACUCUGAUUCCAGGGCAUCCAUCGACA ------..(((.....)-))(((((((((...((((...))))...)))))))))..-..........(((((((.((((......))))....(((((....))))).))))))).... ( -33.60) >DroPer_CAF1 15502 119 - 1 GCAUGCAUCUGGGGCACGAUGGCGGCGGCCAGGUGCUGGGCACCCUCCUCCUGUCGC-CCAUCAAAGAGAUGGACAGCUCCUCAUCGAAUAACACUCUGAUUCCAGGACACCCAGCCACA ........(((((...(((((((((((((.(((.(..((....))..).))))))))-)((((.....))))....))....)))))........((((....))))...)))))..... ( -38.80) >consensus ________CUGGUGCACCAGGACAGCGGCCAGGUGCUGGGCACACUGCUGCUGUCGC_CCAUCAAAGAGAUGGAUAGCUCAUCAUCCAAUAACACUCUGAUUCCAGGGCACCCAGCCACA .............((.....((((((((.(((((((...)))).)))))))))))...(((((.....)))))...)).................((((....))))............. (-26.86 = -26.03 + -0.83)
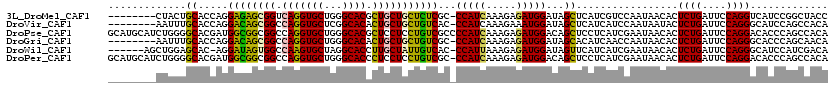

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:34 2006