
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,191,903 – 6,192,002 |
| Length | 99 |
| Max. P | 0.868954 |

| Location | 6,191,903 – 6,192,002 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -19.38 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
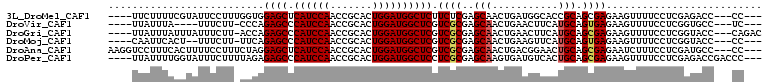
>3L_DroMel_CAF1 6191903 99 + 23771897 ----UUCUUUUCGUAUUCCUUUGGUGGAGCUCAUCCAACCGCACUGGAUGGCUCUUCUCGAGCAACUGAUGGCACCGCAGCGAGAAGUUUUCCUCGAGACC---CC--- ----..................((((((((.((((((.......))))))))))).((((((.((((..(.((......)).)..))))...)))))))))---..--- ( -31.60) >DroVir_CAF1 3408 94 + 1 ----UUAUUUA----UUUCUU-CCCAGAGCCCAUCCAACCGCACUGGAUGGCUCGUCGCGAGCAACUGAACUUCAUGCAGUGAGAAGUUUUCCUCGGUGCC---UC--- ----.......----......-..(((....((((((.......))))))(((((...)))))..)))........(((.((((........)))).))).---..--- ( -24.60) >DroGri_CAF1 3225 101 + 1 ----UUAUUUAUUUAUUUCUU-ACCAGAGCCCAUCCAACCGCACUGGAUGGCUCGUCGCGAGCAACUGAACUUCAUGCAGCGAGAAGUUUUCCUCGGUACC---CAGAC ----.............((((-(((.(((((.(((((.......)))))))))).((((..(((..((.....))))).))))............))))..---.))). ( -27.60) >DroMoj_CAF1 3212 96 + 1 ----CAAUUCACU--UUUCUU-UUCAGAGCCCAUCCAACCGCACUGGAUGGCUCGUCGCGAGCAACUGAAGUUCAUGCAGUGAGAAGUUUUCCUCGGUACC---CC--- ----.........--...(((-(((((((((.(((((.......))))))))))...((((((.......))))..))..)))))))..............---..--- ( -24.60) >DroAna_CAF1 2963 103 + 1 AAGGUCCUUUCACUUUUCCUUUCUAGGAGCUCAUCCAACCGCACUGGAUGGCUCGUCGCGAGCAACUGACGGAACUGCAGCGAGAAUCUUUCCUCGAUGCC---CC--- ..((...........((((.......((((.((((((.......))))))))))((((........))))))))..(((.((((........)))).))))---).--- ( -29.60) >DroPer_CAF1 2705 102 + 1 ----UUAUUUUGGUAUUUCUUUUAGAGAGCCCAUCCAACCGCACUGGAUGGCUCCUCGCGAGCAAGUGAUGUCACUGCAGCGAGAAGUUUUCCUCGAGACCGACCC--- ----.......(((...((((...(((((((((((((.......)))))))...(((((..(((.(((....)))))).)))))..))))))...))))...))).--- ( -35.80) >consensus ____UUAUUUACGUAUUUCUU_CCCAGAGCCCAUCCAACCGCACUGGAUGGCUCGUCGCGAGCAACUGAAGUUAAUGCAGCGAGAAGUUUUCCUCGAUACC___CC___ ..........................((((.((((((.......)))))))))).((((..(((...........))).)))).......................... (-19.38 = -19.55 + 0.17)

| Location | 6,191,903 – 6,192,002 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -24.86 |
| Energy contribution | -25.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6191903 99 - 23771897 ---GG---GGUCUCGAGGAAAACUUCUCGCUGCGGUGCCAUCAGUUGCUCGAGAAGAGCCAUCCAGUGCGGUUGGAUGAGCUCCACCAAAGGAAUACGAAAAGAA---- ---.(---(.(((((((...((((....((......))....)))).))))))).(((((((((((.....))))))).))))..))..................---- ( -31.20) >DroVir_CAF1 3408 94 - 1 ---GA---GGCACCGAGGAAAACUUCUCACUGCAUGAAGUUCAGUUGCUCGCGACGAGCCAUCCAGUGCGGUUGGAUGGGCUCUGGG-AAGAAA----UAAAUAA---- ---..---.(((..(((((....)))))..)))......(((.((((....))))(((((((((((.....)))))).)))))....-..))).----.......---- ( -27.30) >DroGri_CAF1 3225 101 - 1 GUCUG---GGUACCGAGGAAAACUUCUCGCUGCAUGAAGUUCAGUUGCUCGCGACGAGCCAUCCAGUGCGGUUGGAUGGGCUCUGGU-AAGAAAUAAAUAAAUAA---- .((..---.(((.((((((....)))))).)))..))..(((.((((....))))(((((((((((.....)))))).)))))....-..)))............---- ( -29.40) >DroMoj_CAF1 3212 96 - 1 ---GG---GGUACCGAGGAAAACUUCUCACUGCAUGAACUUCAGUUGCUCGCGACGAGCCAUCCAGUGCGGUUGGAUGGGCUCUGAA-AAGAAA--AGUGAAUUG---- ---((---....))((((....))))(((((........((((((((....))))(((((((((((.....)))))).)))))))))-......--)))))....---- ( -29.04) >DroAna_CAF1 2963 103 - 1 ---GG---GGCAUCGAGGAAAGAUUCUCGCUGCAGUUCCGUCAGUUGCUCGCGACGAGCCAUCCAGUGCGGUUGGAUGAGCUCCUAGAAAGGAAAAGUGAAAGGACCUU ---((---((((.((((((....)))))).)))..((((....((((....))))(((((((((((.....))))))).)))).......))))...........))). ( -35.60) >DroPer_CAF1 2705 102 - 1 ---GGGUCGGUCUCGAGGAAAACUUCUCGCUGCAGUGACAUCACUUGCUCGCGAGGAGCCAUCCAGUGCGGUUGGAUGGGCUCUCUAAAAGAAAUACCAAAAUAA---- ---.(((...(((.((((....((((((((.((((((....))).)))..))))))))((((((((.....))))))))..))))....)))...))).......---- ( -37.40) >consensus ___GG___GGUACCGAGGAAAACUUCUCGCUGCAGGAACUUCAGUUGCUCGCGACGAGCCAUCCAGUGCGGUUGGAUGGGCUCUGAA_AAGAAAUAAGUAAAUAA____ .........(((.((((((....)))))).))).......((.((((....))))(((((((((((.....))))))).)))).......))................. (-24.86 = -25.20 + 0.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:27 2006