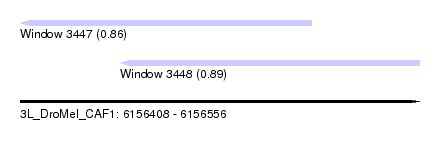
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,156,408 – 6,156,556 |
| Length | 148 |
| Max. P | 0.890790 |

| Location | 6,156,408 – 6,156,516 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 88.27 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -32.83 |
| Energy contribution | -34.67 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6156408 108 - 23771897 AAGGCGGAGGCGGAGCUCCGGAAGAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGUCGAACACUUACGACGGCUUCUACGCGCAGAAGGCGGAG ....(((((((((..((((((.....)).)))).))))))))).((((((.....))))))((((((.((((........)))).)))))).(((.......)))... ( -50.90) >DroSec_CAF1 7805 108 - 1 AAGGCGGAGGCGGAGCUCCGGAAGAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGACGAACACGUACGACGGCUUCUACGCGCAGAAGGCGGAG ....(((((((((..((((((.....)).)))).))))))))).((((((.....))))))((((((.((((....))).)....)))))).(((.......)))... ( -46.40) >DroSim_CAF1 7844 108 - 1 AAGGCGGAGGCGGAGCUCCGGAAGAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGACGAACACGUACGACGGCUUCUACGCGCAGAAAGCGGAG ....(((((((((..((((((.....)).)))).))))))))).((((((.....))))))((((((.((((....))).)....)))))).(((.......)))... ( -47.00) >DroEre_CAF1 7958 108 - 1 AAGGCAGAGGCGGAGCUUCGGAAAAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGACAAACACGUACGAUGGCUUCUACGCUCAGAAGGCGGAG ....(..((((((..((((((.....)).)))).))))))..).((((((.....))))))((((((.(((......)).)....)))))).((((.....))))... ( -37.90) >DroWil_CAF1 8673 108 - 1 AAGGCUGAAAUUGAGCUUAAGAAGAUCCAAGAGGCGGCCUUCGAGCAGCUAAAUAAGCUGCAAAAGCAACAGAAUACCUACGAUGGCUUCUAUGCCCAAAAGAACGCC .(((((.......)))))..............((((..(((...((((((.....))))))....(((..((((..((......)).)))).)))....)))..)))) ( -26.60) >DroYak_CAF1 7961 108 - 1 AAGGCGGAGGCGGAGCUUCGCAAGAUCCAGGAGGCUGCCUUCGAGCAGCUGAACAAGCUGCAGAAGCAGACAAACACGUACGAUGGCUUCUACGCUCAAAAGGCCGAG ..(((((((((((..((((..........)))).))))))))((((((((.....))))(.((((((.(((......)).)....)))))).))))).....)))... ( -41.10) >consensus AAGGCGGAGGCGGAGCUCCGGAAGAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGACGAACACGUACGACGGCUUCUACGCGCAGAAGGCGGAG ....(((((((((..((((((.....)).)))).))))))))).((((((.....))))))((((((...((........))...)))))).(((.......)))... (-32.83 = -34.67 + 1.84)



| Location | 6,156,445 – 6,156,556 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -29.00 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6156445 111 - 23771897 GCUGCUGGCCUACCAGAUCGAGGAGAACGAGGCCAAGCGCAAGGCGGAGGCGGAGCUCCGGAAGAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGUC--------- (((((((((((....(.((.....)).).)))))...(....).(((((((((..((((((.....)).)))).))))))))).((((((.....))))))...)))))).--------- ( -52.00) >DroGri_CAF1 5440 120 - 1 AUUGCUGGCACAACAGAUGGAGGAGAAUGAAGCGAAGCGCAAGGCAGAGGAGGAGACGCGCAAGAUUCAAGAGGCUGCAUGCGAGCAGCUCAACAGACUUCAAAAGCAGGUGCAGAGUGA .(..((.((((......(((((.....(((.((....((((..((((.....(((...(....).)))......)))).))))....))))).....))))).......))))..))..) ( -32.52) >DroSim_CAF1 7881 111 - 1 GCUGCUGGCCUACCAGAUCGAGGAGAACGAGGCCAAGCGGAAGGCGGAGGCGGAGCUCCGGAAGAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGAC--------- .((((((((((....(.((.....)).).))))).)))))...((((((((((..((((((.....)).)))).))))))))..((((((.....))))))....))....--------- ( -51.60) >DroEre_CAF1 7995 111 - 1 GCUGCUGGCCUACCAAAUCGAGGAGAACGAGGCCAAGCGCAAGGCAGAGGCGGAGCUUCGGAAAAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGAC--------- ((.((((((((......((.....))...))))).)))))...((.(((((((..((((((.....)).)))).)))))))...((((((.....))))))....))....--------- ( -42.70) >DroWil_CAF1 8710 111 - 1 GCUGUUGGCAUUCCAAAUCGAAGAGAAUGAGGCUAAACGCAAGGCUGAAAUUGAGCUUAAGAAGAUCCAAGAGGCGGCCUUCGAGCAGCUAAAUAAGCUGCAAAAGCAACA--------- ((((((..((((((........).))))).)))....((.(((((((...(((..((.....))...)))....))))))))).((((((.....))))))...)))....--------- ( -30.70) >DroYak_CAF1 7998 111 - 1 ACUGCUGGCCUACCAGAUCGAGGAGAACGAGGCCAAGCGCAAGGCGGAGGCGGAGCUUCGCAAGAUCCAGGAGGCUGCCUUCGAGCAGCUGAACAAGCUGCAGAAGCAGAC--------- .((((((((((....(.((.....)).).)))))...(....).(((((((((..((((..........)))).))))))))).((((((.....))))))...)))))..--------- ( -46.80) >consensus GCUGCUGGCCUACCAGAUCGAGGAGAACGAGGCCAAGCGCAAGGCGGAGGCGGAGCUUCGGAAGAUCCAGGAGGCUGCCUUCGAGCAGCUAAACAAGCUGCAGAAGCAGAC_________ .((((((((((....(.((.....)).).)))))...((.(((((((....(((....(....).)))......))))))))).((((((.....))))))...)))))........... (-29.00 = -29.98 + 0.98)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:17 2006