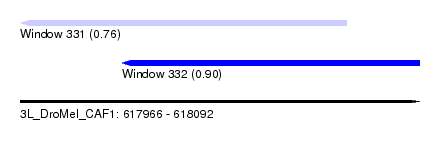
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 617,966 – 618,092 |
| Length | 126 |
| Max. P | 0.901657 |

| Location | 617,966 – 618,069 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.91 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
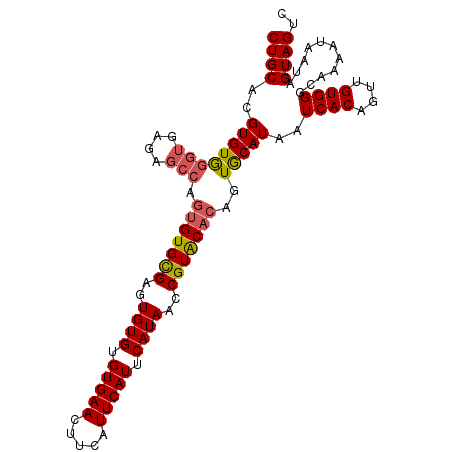
>3L_DroMel_CAF1 617966 103 - 23771897 CUGCACGUGUGGGUGAGUGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCAUAAUCACAGUUGUGGGCAAAAUAAUAGUAGUC (..((..(((((....(((((.(((((((..((((.(((((.....))))).))))..))))))).).))))..)))))..))..)................. ( -31.80) >DroSec_CAF1 29245 103 - 1 CUGCACGUGUAGGUGAGAGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCAUAAUCACAGUUGUGGGCAAAAUAAUAGUAGUC .(((((.....(((....))).(((((((..((((.(((((.....))))).))))..))))))).)))))...((((....))))................. ( -33.70) >DroSim_CAF1 27659 103 - 1 CUGCACGUGUGGGUGAGAGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCAUAAUCACAGUUGUGGGCAAAAUAAUAGUAGUC .(((((.....(((....))).(((((((..((((.(((((.....))))).))))..))))))).)))))...((((....))))................. ( -33.50) >DroEre_CAF1 31533 99 - 1 CUGCACGUGUGGGUG----AGCCAGUGUGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUGCAACGUACAUAAUCACAGUUGUGGCCAAAAUAAUAGUAGUC .((((((.((.((..----..))..((((((((.(((.......))).))))))))))))))))..(..((((((....))))))..)............... ( -25.50) >consensus CUGCACGUGUGGGUGAGAGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCAUAAUCACAGUUGUGGGCAAAAUAAUAGUAGUC ((((..((((((((....))).(((((((..((((.(((((.....))))).))))..)))))))..)))))..((((....))))...........)))).. (-25.25 = -25.75 + 0.50)

| Location | 617,998 – 618,092 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -24.75 |
| Energy contribution | -25.62 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
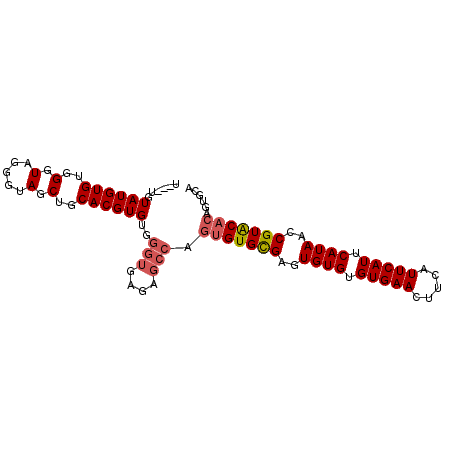
>3L_DroMel_CAF1 617998 94 - 23771897 UGUGGGUAUGUGUGGGUAG-CUAGCUGCACGUGUGGGUGAGUGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCA .....((((.....((((.-.((.(..(....)..).))..)))).(((((((..((((.(((((.....))))).))))..))))))).)))). ( -30.10) >DroSec_CAF1 29277 92 - 1 U---UGUAUGUGUGGGUAGGGUAGCUGCACGUGUAGGUGAGAGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCA .---...(((((..(.((...)).)..)))))((((((....))).(((((((..((((.(((((.....))))).))))..)))))))..))). ( -32.90) >DroSim_CAF1 27691 92 - 1 U---UGUAUGUGUGGGUAGGGUAGCUGCACGUGUGGGUGAGAGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCA .---..((((((..(.((...)).)..))))))..(((....))).(((((((..((((.(((((.....))))).))))..)))))))...... ( -32.50) >DroEre_CAF1 31565 87 - 1 U---UGUAUGUGUGGGU-GGGUAGCUGCACGUGUGGGUG----AGCCAGUGUGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUGCAACGUACA .---(((((((...(((-.....)))(((((.((.((..----..))..((((((((.(((.......))).))))))))))))))).))))))) ( -27.10) >consensus U___UGUAUGUGUGGGUAGGGUAGCUGCACGUGUGGGUGAGAGCCAGUGUGCGAGUGUGUGUGAACUUCAUUCAUUCAUAACCGUACACAGUGCA ......((((((..(.(.....).)..))))))..(((....))).(((((((..((((.(((((.....))))).))))..)))))))...... (-24.75 = -25.62 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:45 2006