
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
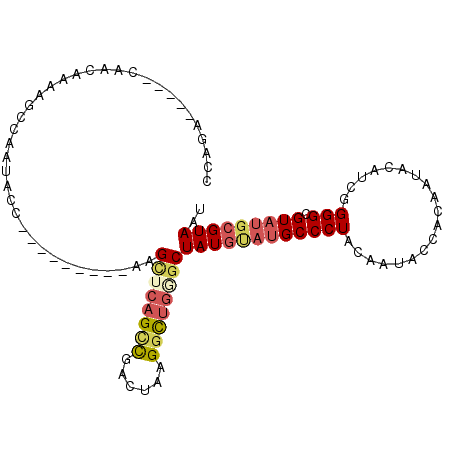
| Location | 6,114,955 – 6,115,071 |
| Length | 116 |
| Max. P | 0.977572 |

| Location | 6,114,955 – 6,115,047 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -20.15 |
| Energy contribution | -21.01 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6114955 92 + 23771897 CCAGA-----CAACAAAAGCCAAUACC---------AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACCACAAUACAUCGGGGCGUAUGCGUAAU .....-----........((.......---------.(((((((((......)))))))))..(((((((((..................)))))))))))..... ( -29.97) >DroSec_CAF1 20548 86 + 1 CCAGA-----CAACAAAAGCC---------------AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACAGCAAUACAUCGGGGCGUAUGCGUAAU .....-----........((.---------------.(((((((((......)))))))))..(((((((((..................)))))))))))..... ( -29.97) >DroSim_CAF1 21230 86 + 1 CCAGA-----CAACAAAAGCC---------------AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACAACAAUACAUCGGGGCGUAUGCGUAAU .....-----........((.---------------.(((((((((......)))))))))..(((((((((..................)))))))))))..... ( -29.97) >DroEre_CAF1 24201 92 + 1 CCAGA-----CAACAAAAGCCAAUACC---------AAGCUCAGUCGACUAUGGCUGAGCUAUGCAUGCCCUACAAUACCACAAUACAACUGGGCGUAUGCGUAAU .....-----.................---------..((((((((......))))))))(((((((((((((.................)))).))))))))).. ( -26.83) >DroYak_CAF1 23678 92 + 1 CCAGA-----CAACAAAAGCCAAUACC---------AAGCUCAGUCGACUAUGGCUCUACUAUGCAUGCCCUACAAUACCACAAUACAACUGGGCGUAUGCGUAAU .....-----.......(((((.....---------...............)))))....(((((((((((((.................)))).))))))))).. ( -17.48) >DroPer_CAF1 29213 95 + 1 GCAGAAGCGGCAACAACAGCCAAUAUCUGACACUUUCAGUUCAG-UGUCUGCGGUCGUGCUAU----GCCCUACAAUAAC-----CCAUC-GGGCGUAUGAGUAAU (((((.(((((.......))).....((((.....))))....)-).)))))(.((((((...----((((.........-----.....-)))))))))).)... ( -26.94) >consensus CCAGA_____CAACAAAAGCCAAUACC_________AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACCACAAUACAUCGGGGCGUAUGCGUAAU ......................................((((((((......))))))))((((((((((((...................))).))))))))).. (-20.15 = -21.01 + 0.86)


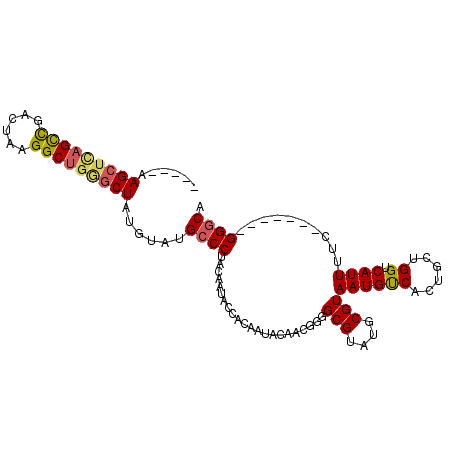
| Location | 6,114,977 – 6,115,071 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -23.37 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6114977 94 + 23771897 -----AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACCACAAUACAUCGGGGCGUAUGCGUAAUGUCACUGCUGGUCAUUUUC-------GGGCA -----.(((((((((......))))))))).....(((((...(((((((((..((((....(((....))).))))...)).)))).)))...-------))))) ( -33.70) >DroSec_CAF1 20564 94 + 1 -----AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACAGCAAUACAUCGGGGCGUAUGCGUAAUGUCACUGCUGGUCAUUUUC-------GGGCA -----.(((((((((......))))))))).....(((((.......(((((..((((....(((....))).))))...))))).........-------))))) ( -35.59) >DroSim_CAF1 21246 94 + 1 -----AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACAACAAUACAUCGGGGCGUAUGCGUAAUGUCACUGCUGGUCAUUUUC-------GGGCA -----.(((((((((......)))))))))..(((((((((..................))))))))).............(((.(.......)-------.))). ( -32.27) >DroEre_CAF1 24223 94 + 1 -----AAGCUCAGUCGACUAUGGCUGAGCUAUGCAUGCCCUACAAUACCACAAUACAACUGGGCGUAUGCGUAAUGUCACUGCUGGUCAUUUUC-------GGGCA -----..((((((((......))))))))(((((((((((((.................)))).)))))))))........(((.(.......)-------.))). ( -30.63) >DroYak_CAF1 23700 94 + 1 -----AAGCUCAGUCGACUAUGGCUCUACUAUGCAUGCCCUACAAUACCACAAUACAACUGGGCGUAUGCGUAAUGUCACUGCUGGUCAUUUUC-------GGGCG -----..((((((..(((((((((.....(((((((((((((.................)))).)))))))))..))))....)))))...)).-------)))). ( -24.23) >DroAna_CAF1 28980 94 + 1 GAUGCGAGCUUCGCUAACAUCGGCUGGGCUAU----GCCCUACAAU--------ACAACGGGGCGUAUGCGUAAUGCCAUGUCAGGUCAUUUGCCGGGCUGGGCCA ((((..(((...)))..))))((((.((((((----(((((.....--------.....)))))))..(((.((((((......)).)))))))..)))).)))). ( -36.40) >consensus _____AAGCUCAGCCGACUAAGGCUGGGCUAUGUAUGCCCUACAAUACCACAAUACAACGGGGCGUAUGCGUAAUGUCACUGCUGGUCAUUUUC_______GGGCA ......(((((((((......)))))))))......((((......................(((....)))((((((......)).))))..........)))). (-23.37 = -23.48 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:08 2006