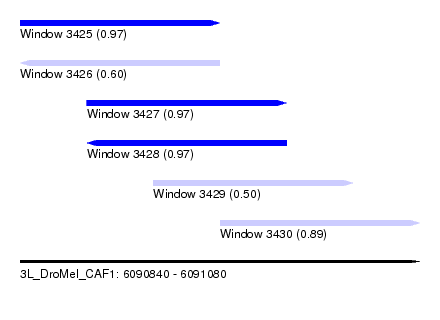
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,090,840 – 6,091,080 |
| Length | 240 |
| Max. P | 0.974331 |

| Location | 6,090,840 – 6,090,960 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6090840 120 + 23771897 UUUACUAAAUUUUCGCUGAAACGAAAGUCACUUUCAAAAUGGUCGGAUUCAAUGCCUCUGACCAGUUGCCAAGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGAC ........(((((((......))))))).....((((((((((((((.........)))))))).(((((..(((..(((((((.....)))))))..))).....)))))..)))))). ( -33.40) >DroSec_CAF1 39189 120 + 1 UUUACUACAUUUUCGCUGAAACGAAAGUCACUUUCAAAAUGGUCGGAUUCAAUGCCUCUGACCAGUUGCCAGGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGAC ........(((((((......))))))).....((((((((((((((.........))))))))(((.(((((((..(((((((.....)))))))..))).)))))))....)))))). ( -34.30) >DroSim_CAF1 40509 120 + 1 UUUACUACAUUUUCGCUGAAAUGAAAGUCACUUUCAAAAUGGUCGGAUUCAAUGCCUCUGACCAGUUGCCAGGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGAC ..............(((....((((((...))))))...((((((((.........))))))))(((.(((((((..(((((((.....)))))))..))).))))))).)))....... ( -32.90) >consensus UUUACUACAUUUUCGCUGAAACGAAAGUCACUUUCAAAAUGGUCGGAUUCAAUGCCUCUGACCAGUUGCCAGGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGAC ........(((((((......))))))).....((((((((((((((.........)))))))).(((((..(((..(((((((.....)))))))..))).....)))))..)))))). (-32.22 = -32.00 + -0.22)

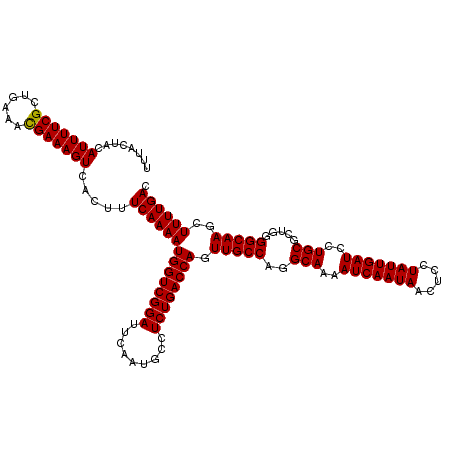
| Location | 6,090,840 – 6,090,960 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -32.25 |
| Energy contribution | -32.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6090840 120 - 23771897 GUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCUUGGCAACUGGUCAGAGGCAUUGAAUCCGACCAUUUUGAAAGUGACUUUCGUUUCAGCGAAAAUUUAGUAAA ((((..((.(((((.....(((((((((((((...)))))))))))))..)))))))((((.((..(...)..)).))))..........))))((((((....)))))).......... ( -32.90) >DroSec_CAF1 39189 120 - 1 GUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCCUGGCAACUGGUCAGAGGCAUUGAAUCCGACCAUUUUGAAAGUGACUUUCGUUUCAGCGAAAAUGUAGUAAA ((((..((.(((((..((.(((((((((((((...))))))))))))))))))))))((((.((..(...)..)).))))..........))))((((((....)))))).......... ( -34.90) >DroSim_CAF1 40509 120 - 1 GUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCCUGGCAACUGGUCAGAGGCAUUGAAUCCGACCAUUUUGAAAGUGACUUUCAUUUCAGCGAAAAUGUAGUAAA .((((((..(((((..((.(((((((((((((...)))))))))))))))))))).(((((.((..(...)..)).))))))))))).....(((..(((((......))))).)))... ( -32.70) >consensus GUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCCUGGCAACUGGUCAGAGGCAUUGAAUCCGACCAUUUUGAAAGUGACUUUCGUUUCAGCGAAAAUGUAGUAAA ((((..((.(((((..((.(((((((((((((...))))))))))))))))))))))((((.((..(...)..)).))))..........))))((((((....)))))).......... (-32.25 = -32.37 + 0.11)

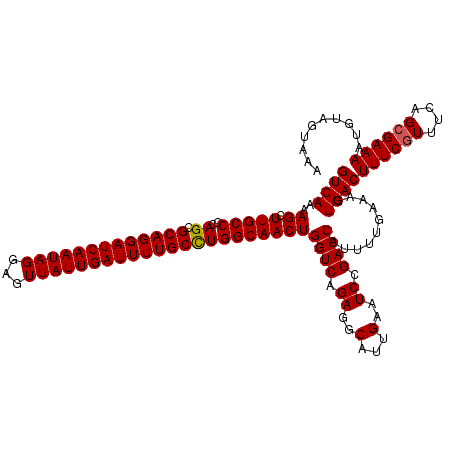
| Location | 6,090,880 – 6,091,000 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -40.27 |
| Energy contribution | -40.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6090880 120 + 23771897 GGUCGGAUUCAAUGCCUCUGACCAGUUGCCAAGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCC (((((((.........))))))).....(((.(((..(((((((.....)))))))..)))..)))(((((.....((..((((((.....))))))..)).......)))))....... ( -39.40) >DroSec_CAF1 39229 120 + 1 GGUCGGAUUCAAUGCCUCUGACCAGUUGCCAGGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUAUCC (((((((.........))))))).....(((((((..(((((((.....)))))))..))).))))(((((.....((..((((((.....))))))..)).......)))))....... ( -42.40) >DroSim_CAF1 40549 120 + 1 GGUCGGAUUCAAUGCCUCUGACCAGUUGCCAGGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCC (((((((.........))))))).....(((((((..(((((((.....)))))))..))).))))(((((.....((..((((((.....))))))..)).......)))))....... ( -42.40) >consensus GGUCGGAUUCAAUGCCUCUGACCAGUUGCCAGGCAAAAUCAAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCC (((((((.........))))))).....(((((((..(((((((.....)))))))..))).))))(((((.....((..((((((.....))))))..)).......)))))....... (-40.27 = -40.60 + 0.33)


| Location | 6,090,880 – 6,091,000 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -45.60 |
| Consensus MFE | -46.01 |
| Energy contribution | -45.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6090880 120 - 23771897 GGACAAUGGCAACGGUGGGUGGAGCCACCAUAAUGGCGGCGUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCUUGGCAACUGGUCAGAGGCAUUGAAUCCGACC ((((((((.(...((..(((.(((((.((.....)).))).))....)))..))((((.(((((((((((((...)))))))))))))))))............).)))))..))).... ( -44.30) >DroSec_CAF1 39229 120 - 1 GGAUAAUGGCAACGGUGGGUGGAGCCACCAUAAUGGCGGCGUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCCUGGCAACUGGUCAGAGGCAUUGAAUCCGACC ((((((((.(...((..(((.(((((.((.....)).))).))....)))..))((((.(((((((((((((...)))))))))))))))))............).))))..)))).... ( -45.50) >DroSim_CAF1 40549 120 - 1 GGACAAUGGCAACGGUGGGUGGAGCCACCAUAAUGGCGGCGUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCCUGGCAACUGGUCAGAGGCAUUGAAUCCGACC ((((((((.(...((..(((.(((((.((.....)).))).))....)))..))((((.(((((((((((((...)))))))))))))))))............).)))))..))).... ( -47.00) >consensus GGACAAUGGCAACGGUGGGUGGAGCCACCAUAAUGGCGGCGUCAAAAGCUUGCCCCAGCGCAGGAUCAAUAGGAGUUAUUGAUUUUGCCUGGCAACUGGUCAGAGGCAUUGAAUCCGACC ((((((((.(...((..(((.(((((.((.....)).))).))....)))..))((((.(((((((((((((...)))))))))))))))))............).)))))..))).... (-46.01 = -45.57 + -0.44)

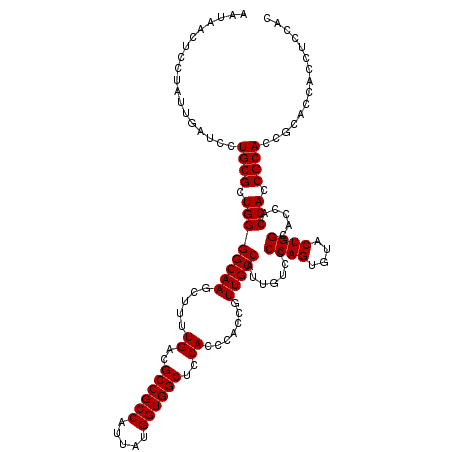
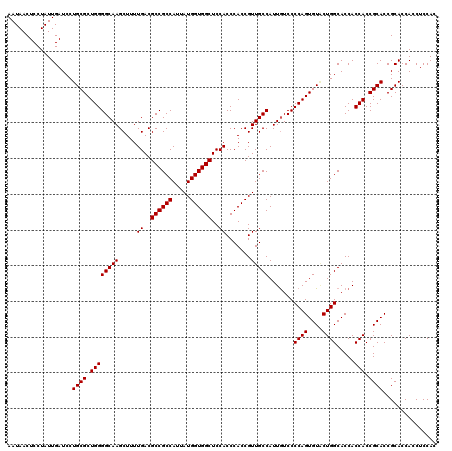
| Location | 6,090,920 – 6,091,040 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -31.37 |
| Energy contribution | -31.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6090920 120 + 23771897 AAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCCCCAGUGUGCUGGCACCACCACCGCACCGCACCACCUCCAC ..................(((((((((((((((.......)).((....(((((((......))))))).))..))).)))))))(((((((.....)))..)))).))).......... ( -34.90) >DroSec_CAF1 39269 120 + 1 AAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUAUCCCCAGUGUACUGGCACCACCACCGCACCGCACCACCUCCAC ..................((((.((((((((.....((..((((((.....))))))..)).......))))).......((((....)))).....))).))))............... ( -31.40) >DroSim_CAF1 40589 120 + 1 AAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCCCCAGUGUUCUGGCACCACCACCGCACCGCACCACCUCCAC ..................((((.((((((((((.......)).((....(((((((......))))))).))..))).)))))((((..(((.....)))..)))))))).......... ( -31.40) >consensus AAUAACUCCUAUUGAUCCUGCGCUGGGGCAAGCUUUUGACGCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCCCCAGUGUACUGGCACCACCACCGCACCGCACCACCUCCAC ..................((((.((((((((.....((..((((((.....))))))..)).......))))).......((((....)))).....))).))))............... (-31.37 = -31.37 + -0.00)

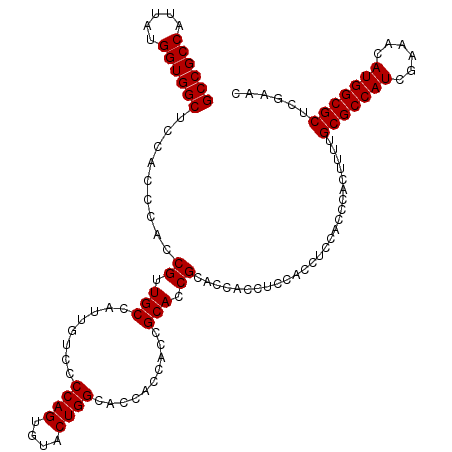
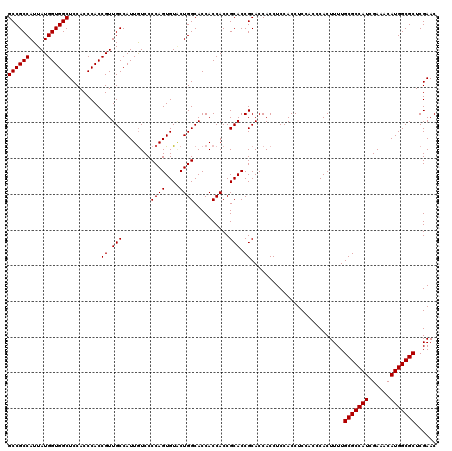
| Location | 6,090,960 – 6,091,080 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -30.04 |
| Energy contribution | -30.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6090960 120 + 23771897 GCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCCCCAGUGUGCUGGCACCACCACCGCACCGCACCACCUCCACCUCUACCCACUUUUGCGCCAUCGAAACAUGGCGCUCGAAC ((((((.....))))))..........(.(((...((....))(((((.(((.....))).))))).))).)......................(((((((......)))))))...... ( -32.80) >DroSec_CAF1 39309 120 + 1 GCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUAUCCCCAGUGUACUGGCACCACCACCGCACCGCACCACCUCCACCUCCACCCACUUUUGCGCCAUCGAAACAUGGCGCUCGAAC ((((((.....)))))).........((.(((........((((....))))..........))).))..........................(((((((......)))))))...... ( -30.07) >DroSim_CAF1 40629 120 + 1 GCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCCCCAGUGUUCUGGCACCACCACCGCACCGCACCACCUCCACCUCCACCCACUUUUGCGCCAUCGAAACAUGGCGCUCGAAC ((((((.....)))))).........((.((((((((....)))))...(((.....)))..))).))..........................(((((((......)))))))...... ( -30.40) >consensus GCCGCCAUUAUGGUGGCUCCACCCACCGUUGCCAUUGUCCCCAGUGUACUGGCACCACCACCGCACCGCACCACCUCCACCUCCACCCACUUUUGCGCCAUCGAAACAUGGCGCUCGAAC ((((((.....)))))).........((.(((........((((....))))..........))).))..........................(((((((......)))))))...... (-30.04 = -30.04 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:59 2006