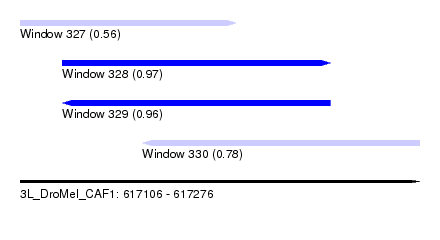
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 617,106 – 617,276 |
| Length | 170 |
| Max. P | 0.968351 |

| Location | 617,106 – 617,198 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -19.56 |
| Energy contribution | -21.08 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 617106 92 + 23771897 CGUCGGCGUCUCGUAUCAUCGCUC------CAACAACAACAAAAACGGCAGCACUAUGGCAGCAGCAACCUCAAAUGCGCUCUUGCUACUGCUGCCGA .((.((((...........)))).------..))...........((((((((...((((((.(((............))).)))))).)))))))). ( -27.00) >DroSec_CAF1 28400 89 + 1 CGUCGGCGUCUCGUAUCAUCGCUC------CAACA---ACAAAAGCGGCAGCACUACGGCAGCAGCAACCUCAAAUGCGCUCUUGCUGCUGCUGCCGA ..(((((....((......))...------.....---.....((((((((((....((.(((.(((........))))))))))))))))))))))) ( -30.30) >DroSim_CAF1 26790 89 + 1 CGUCGGCGUCUCGUAUCAUCGCUC------CAACA---ACAAAAGCGGCAGCACUAUGGCAGCAGCAACCUCAAAUGCGCUCUUGCUGCUGCUGCCGA ..(((((....((......))...------.....---.....((((((((((....((.(((.(((........))))))))))))))))))))))) ( -30.30) >DroEre_CAF1 30509 77 + 1 CGCCGGCGUCCCGUAUCAUCGCU------ACAACA---ACAAAAGCGGCAGCACUGUGGCAGCAGCAACCUCAAAUGCGCUCA------------CGC .(((((....))........(((------......---.....))))))......(((((.(((...........))))).))------------).. ( -15.40) >DroYak_CAF1 32166 94 + 1 CGUCGGCGUCUCGUAUCAUCGCUCCACAAACAACA---ACAAAAGCGGCGGCACUGUGGCAGCAGCAACCUCGAAUGCGCUCU-GCCGCUGCUGCCGA .((.((((...........))).).))........---.......((((((((..(((((((..(((........)))...))-))))))))))))). ( -33.20) >consensus CGUCGGCGUCUCGUAUCAUCGCUC______CAACA___ACAAAAGCGGCAGCACUAUGGCAGCAGCAACCUCAAAUGCGCUCUUGCUGCUGCUGCCGA ....((((...........))))......................((((((((...(((((((.(((........))))))...)))).)))))))). (-19.56 = -21.08 + 1.52)

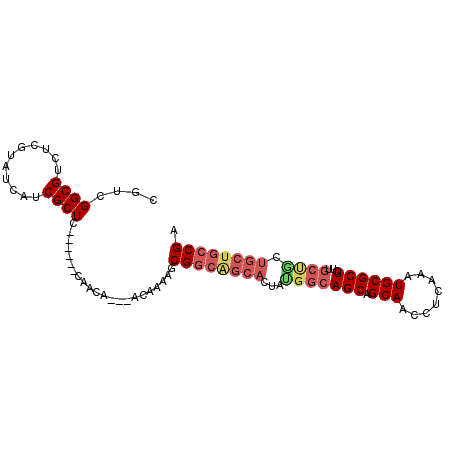
| Location | 617,124 – 617,238 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -16.88 |
| Energy contribution | -18.40 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 617124 114 + 23771897 UCGCUC------CAACAACAACAAAAACGGCAGCACUAUGGCAGCAGCAACCUCAAAUGCGCUCUUGCUACUGCUGCCGAUGCACUGCAAGAAAUGGACGCCAAAUAUGAAAUAUCCCAA ..((((------((.............((((((((...((((((.(((............))).)))))).)))))))).(((...))).....)))).))................... ( -31.10) >DroSec_CAF1 28418 103 + 1 UCGCUC------CAACA---ACAAAAGCGGCAGCACUACGGCAGCAGCAACCUCAAAUGCGCUCUUGCUGCUGCUGCCGAUGCACUGAAAGAAACGGA--------GUGAAAUAUCCCCU ((((((------(....---......((....))....(((((((((((..(......).((....)))))))))))))................)))--------)))).......... ( -36.10) >DroSim_CAF1 26808 103 + 1 UCGCUC------CAACA---ACAAAAGCGGCAGCACUAUGGCAGCAGCAACCUCAAAUGCGCUCUUGCUGCUGCUGCCGAUGCACUGAAAGAAACGGA--------GUGAAAUAUCCCCU ((((((------(....---.....((((((((((....((.(((.(((........))))))))))))))))))((....))............)))--------)))).......... ( -35.10) >DroEre_CAF1 30527 99 + 1 UCGCU------ACAACA---ACAAAAGCGGCAGCACUGUGGCAGCAGCAACCUCAAAUGCGCUCA------------CGCUGCACUGAGAGAAAUGGACGCAGAACAUAAAAUAUCCCAA .((((------......---.....))))(((((...(((((.(((...........))))).))------------)))))).(((.(.........).)))................. ( -19.10) >DroYak_CAF1 32184 116 + 1 UCGCUCCACAAACAACA---ACAAAAGCGGCGGCACUGUGGCAGCAGCAACCUCGAAUGCGCUCU-GCCGCUGCUGCCGAUGCACUGCAAGAAAUGAACGAAGGACAUAGGAUAUCCCAA ..(.(((..........---.......((((((((..(((((((..(((........)))...))-))))))))))))).(((...))).............))))...((.....)).. ( -34.80) >consensus UCGCUC______CAACA___ACAAAAGCGGCAGCACUAUGGCAGCAGCAACCUCAAAUGCGCUCUUGCUGCUGCUGCCGAUGCACUGAAAGAAAUGGACG____A_AUGAAAUAUCCCAA ...........................((((((((...(((((((.(((........))))))...)))).))))))))......................................... (-16.88 = -18.40 + 1.52)


| Location | 617,124 – 617,238 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -24.59 |
| Energy contribution | -26.87 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 617124 114 - 23771897 UUGGGAUAUUUCAUAUUUGGCGUCCAUUUCUUGCAGUGCAUCGGCAGCAGUAGCAAGAGCGCAUUUGAGGUUGCUGCUGCCAUAGUGCUGCCGUUUUUGUUGUUGUUG------GAGCGA ...((((...(((....))).)))).......((((..(...((((((((((((.(((.....)))...))))))))))))...)..))))((((((..........)------))))). ( -39.40) >DroSec_CAF1 28418 103 - 1 AGGGGAUAUUUCAC--------UCCGUUUCUUUCAGUGCAUCGGCAGCAGCAGCAAGAGCGCAUUUGAGGUUGCUGCUGCCGUAGUGCUGCCGCUUUUGU---UGUUG------GAGCGA .((((........)--------)))........(((..(..(((((((((((((.(((.....)))...)))))))))))))..)..))).((((((...---....)------))))). ( -44.10) >DroSim_CAF1 26808 103 - 1 AGGGGAUAUUUCAC--------UCCGUUUCUUUCAGUGCAUCGGCAGCAGCAGCAAGAGCGCAUUUGAGGUUGCUGCUGCCAUAGUGCUGCCGCUUUUGU---UGUUG------GAGCGA .((((........)--------)))........(((..(...((((((((((((.(((.....)))...))))))))))))...)..))).((((((...---....)------))))). ( -41.20) >DroEre_CAF1 30527 99 - 1 UUGGGAUAUUUUAUGUUCUGCGUCCAUUUCUCUCAGUGCAGCG------------UGAGCGCAUUUGAGGUUGCUGCUGCCACAGUGCUGCCGCUUUUGU---UGUUGU------AGCGA ..(((((((...)))))))...........(((((((((.((.------------...))))).))))))(((((((.((.((((.((....))..))))---.)).))------))))) ( -30.00) >DroYak_CAF1 32184 116 - 1 UUGGGAUAUCCUAUGUCCUUCGUUCAUUUCUUGCAGUGCAUCGGCAGCAGCGGC-AGAGCGCAUUCGAGGUUGCUGCUGCCACAGUGCCGCCGCUUUUGU---UGUUGUUUGUGGAGCGA ..(((((((...)))))))((((((.......((.(..(...((((((((((((-.(((....)))...))))))))))))...)..).))(((......---........))))))))) ( -42.64) >consensus UUGGGAUAUUUCAU_U____CGUCCAUUUCUUUCAGUGCAUCGGCAGCAGCAGCAAGAGCGCAUUUGAGGUUGCUGCUGCCAUAGUGCUGCCGCUUUUGU___UGUUG______GAGCGA ...((((..............))))........(((..(...((((((((((((..(((....)))...))))))))))))...)..))).(((((..................))))). (-24.59 = -26.87 + 2.28)

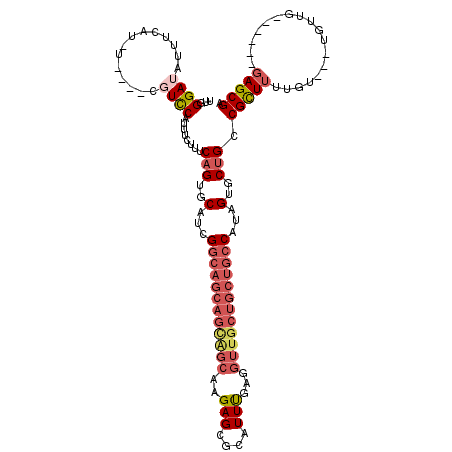
| Location | 617,158 – 617,276 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -16.42 |
| Energy contribution | -19.26 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 617158 118 - 23771897 A-AAUACACGCUA-ACCAGCAAAAACUAUUAUUUCCAGCAUUGGGAUAUUUCAUAUUUGGCGUCCAUUUCUUGCAGUGCAUCGGCAGCAGUAGCAAGAGCGCAUUUGAGGUUGCUGCUGC .-....((((((.-...))).................(((...((((...(((....))).))))......))).))).....(((((((((((.(((.....)))...))))))))))) ( -29.40) >DroSec_CAF1 28449 110 - 1 A-AAUGCACACUA-ACCAGCAAAAACUAUUAUUUCCAGCAAGGGGAUAUUUCAC--------UCCGUUUCUUUCAGUGCAUCGGCAGCAGCAGCAAGAGCGCAUUUGAGGUUGCUGCUGC .-.((((((....-...(((.........(((..((.....))..)))......--------...))).......))))))..(((((((((((.(((.....)))...))))))))))) ( -32.31) >DroSim_CAF1 26839 110 - 1 A-AAUGCACACUA-ACCAGCAAAAACUAUUAUUUCCAGCAAGGGGAUAUUUCAC--------UCCGUUUCUUUCAGUGCAUCGGCAGCAGCAGCAAGAGCGCAUUUGAGGUUGCUGCUGC .-.((((((....-...(((.........(((..((.....))..)))......--------...))).......))))))..(((((((((((.(((.....)))...))))))))))) ( -32.31) >DroEre_CAF1 30558 108 - 1 AAAAUGUACUCUACGCUACCCAAUGCUAUUAUUUUUAGCAUUGGGAUAUUUUAUGUUCUGCGUCCAUUUCUCUCAGUGCAGCG------------UGAGCGCAUUUGAGGUUGCUGCUGC .(((((..(((.(((((.((((((((((.......)))))))))).........((.(((.(.........).))).))))))------------))))..)))))..(((....))).. ( -35.30) >DroYak_CAF1 32221 115 - 1 ----UGUACACUUAGCUACCCAAUACUUUUAUUUUUAGCAUUGGGAUAUCCUAUGUCCUUCGUUCAUUUCUUGCAGUGCAUCGGCAGCAGCGGC-AGAGCGCAUUCGAGGUUGCUGCUGC ----(((((.....((((...((((....))))..))))...(((((((...)))))))................)))))...(((((((((((-.(((....)))...))))))))))) ( -33.00) >consensus A_AAUGCACACUA_ACCAGCAAAAACUAUUAUUUCCAGCAUUGGGAUAUUUCAU_U____CGUCCAUUUCUUUCAGUGCAUCGGCAGCAGCAGCAAGAGCGCAUUUGAGGUUGCUGCUGC .....................................(((((((((........................)))))))))....(((((((((((..(((....)))...))))))))))) (-16.42 = -19.26 + 2.84)

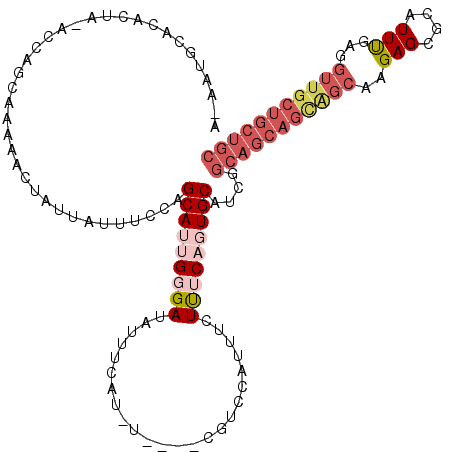
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:44 2006