
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,089,532 – 6,089,754 |
| Length | 222 |
| Max. P | 0.944695 |

| Location | 6,089,532 – 6,089,640 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -24.81 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6089532 108 + 23771897 UCCUUU-UUUCCGAGUGUCGC---UACGCAUACA--------CACACUUACACAGCGCGAGCAGGAAACUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGUGUGUGUG ......-.......((((...---.))))(((((--------(((((((((((.(((((((.......)))))))...........((((((....)))))).)))))))))))))))). ( -42.70) >DroSec_CAF1 37884 115 + 1 AUCUUU-UUUCCGAGUGUCGCAUACACGCAUACACACAAGCACACACUUACACAGC--GAGCAGGAAUCUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU--GUGUG ......-.......((((.((......))..))))....((((((((((((((.((--(((.......))))).............((((((....)))))).))))))))))--)))). ( -37.60) >DroSim_CAF1 39198 116 + 1 AUCUUUUUUUCCGAGUGUCGCAUACACGCAUACACACCAACACACACUUACACAGC--GAGCAGGAAACUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU--GUGUG ..............((((.((......))..))))....((((((((((((((.((--(((.......))))).............((((((....)))))).))))))))))--)))). ( -37.90) >consensus AUCUUU_UUUCCGAGUGUCGCAUACACGCAUACACAC_A_CACACACUUACACAGC__GAGCAGGAAACUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU__GUGUG ..............((((.((......))))))......((((((((((((((.......((((....)).)).............((((((....)))))).))))))))))..)))). (-24.81 = -25.70 + 0.89)



| Location | 6,089,562 – 6,089,680 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -41.53 |
| Consensus MFE | -33.23 |
| Energy contribution | -33.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6089562 118 + 23771897 --CACACUUACACAGCGCGAGCAGGAAACUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGUGUGUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUCUGCA --(((((((((((.(((((((.......)))))))...........((((((....)))))).)))))))))))((((((((..(((....).))..))))))))......((....)). ( -45.00) >DroSec_CAF1 37923 116 + 1 CACACACUUACACAGC--GAGCAGGAAUCUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU--GUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUAUGCA (((((((((((((.((--(((.......))))).............((((((....)))))).))))))))))--)))((((.(((((.........)))))((.......))...)))) ( -39.60) >DroSim_CAF1 39238 116 + 1 CACACACUUACACAGC--GAGCAGGAAACUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU--GUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUCUGCA (((((((((((((.((--(((.......))))).............((((((....)))))).))))))))))--)))((((.(((((.........)))))((.......))...)))) ( -40.00) >consensus CACACACUUACACAGC__GAGCAGGAAACUCGCGCCGGAUAAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU__GUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUCUGCA ...((((((((((.......((((....)).)).............((((((....)))))).))))))))))..(((((((..(((....).))..))))))).......((....)). (-33.23 = -33.23 + -0.00)

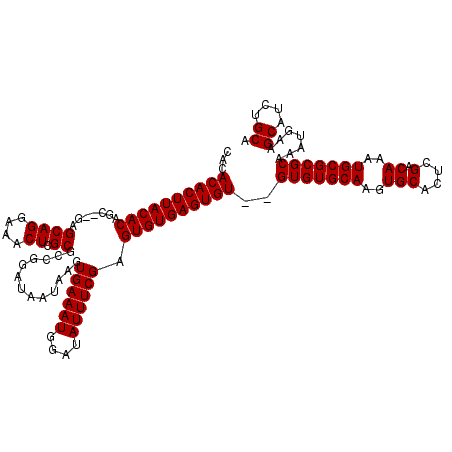

| Location | 6,089,562 – 6,089,680 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -39.97 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6089562 118 - 23771897 UGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACACACACACUCACACUCGAAAUAUCCAUUUCACUUAUUAUCCGGCGCGAGUUUCCUGCUCGCGCUGUGUAAGUGUG-- .((((((((...........))))))))...((((....))))...((((((.((((..(((((....)))))...........(((((((((.....))))))))))))).))))))-- ( -44.50) >DroSec_CAF1 37923 116 - 1 UGCAUAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACAC--ACACUCACACUCGAAAUAUCCAUUUCACUUAUUAUCCGGCGCGAGAUUCCUGCUC--GCUGUGUAAGUGUGUG ((((..(((..((..((....))..))......)))..))))(((--(((((.((((..(((((....)))))..............(((((.......)))--)).)))).)))))))) ( -35.30) >DroSim_CAF1 39238 116 - 1 UGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACAC--ACACUCACACUCGAAAUAUCCAUUUCACUUAUUAUCCGGCGCGAGUUUCCUGCUC--GCUGUGUAAGUGUGUG .(((((((...)))))))..(((..(((.....)))..))).(((--(((((.((((..(((((....)))))..............((((((.....))))--)).)))).)))))))) ( -40.10) >consensus UGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACAC__ACACUCACACUCGAAAUAUCCAUUUCACUUAUUAUCCGGCGCGAGUUUCCUGCUC__GCUGUGUAAGUGUGUG .((((((((...........))))))))...((((....))))....(((((.((((..(((((....)))))...........(((..((((.....))))..))))))).)))))... (-28.67 = -29.33 + 0.67)


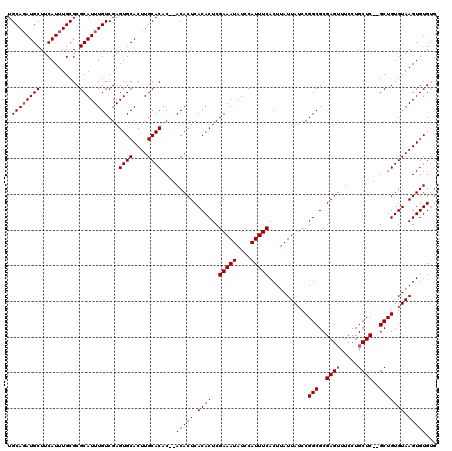
| Location | 6,089,600 – 6,089,714 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -25.73 |
| Energy contribution | -25.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6089600 114 + 23771897 AAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGUGUGUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUCUGCAUGCACACAUA------AGCAAAACAUAUAAAUAUAUACAC ......((((((....))))))..(((..(((((((((((((.(((((.........)))))((.......))...))))))))))))).------.))).................... ( -30.30) >DroSec_CAF1 37961 118 + 1 AAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU--GUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUAUGCAUGCACACAUAUGUAUAUGCAAAACAUAUAAAAAUAUACAC ......((((((....)))))).(((((.((((--(((((((.(((((.........)))))(((.(((...))).))).))))))))))).((((((.....))))))......))))) ( -33.20) >DroSim_CAF1 39276 118 + 1 AAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU--GUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUCUGCAUGCACACAUAUGUAUAUGCAAAACAUAUAAAAAUAUACAC ......((((((....)))))).(((((.((((--(((((((.(((((.........)))))(((.(((...))).))).))))))))))).((((((.....))))))......))))) ( -33.20) >consensus AAUAAGUGAAAUGGAUAUUUCGAGUGUGAGUGU__GUGUGCAAGUGCACUCGACAAAUGCGCGCAAAUGAAGCAUCUGCAUGCACACAUAUGUAUAUGCAAAACAUAUAAAAAUAUACAC .....(((...........(((((((((..(((......)))..)))))))))....((((.(((.(((...))).))).)))))))................................. (-25.73 = -25.73 + -0.00)



| Location | 6,089,600 – 6,089,714 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -25.73 |
| Energy contribution | -27.07 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6089600 114 - 23771897 GUGUAUAUAUUUAUAUGUUUUGCU------UAUGUGUGCAUGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACACACACACUCACACUCGAAAUAUCCAUUUCACUUAUU (((((((((....)))))......------..((((((((.(((((((...)))))))..((((((...))))))...)))))))).))))........(((((....)))))....... ( -28.30) >DroSec_CAF1 37961 118 - 1 GUGUAUAUUUUUAUAUGUUUUGCAUAUACAUAUGUGUGCAUGCAUAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACAC--ACACUCACACUCGAAAUAUCCAUUUCACUUAUU ((((.......((((((.....))))))....((((((((.(((.(((...))).)))..((((((...))))))...)))))))--).....))))..(((((....)))))....... ( -29.30) >DroSim_CAF1 39276 118 - 1 GUGUAUAUUUUUAUAUGUUUUGCAUAUACAUAUGUGUGCAUGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACAC--ACACUCACACUCGAAAUAUCCAUUUCACUUAUU ((((.......((((((.....))))))....((((((((.(((((((...)))))))..((((((...))))))...)))))))--).....))))..(((((....)))))....... ( -32.70) >consensus GUGUAUAUUUUUAUAUGUUUUGCAUAUACAUAUGUGUGCAUGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUGCACAC__ACACUCACACUCGAAAUAUCCAUUUCACUUAUU ((((........((((((.........))))))(((((((.(((((((...)))))))..((((((...))))))...)))))))........))))..(((((....)))))....... (-25.73 = -27.07 + 1.33)

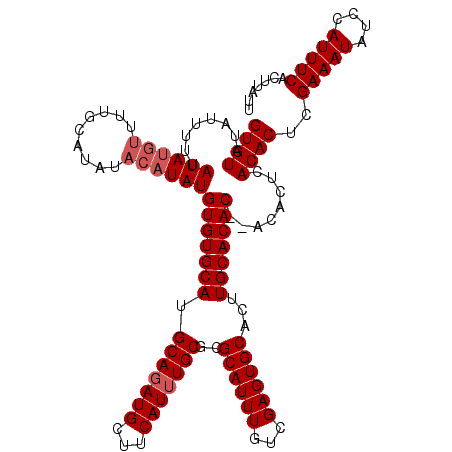

| Location | 6,089,640 – 6,089,754 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -26.23 |
| Energy contribution | -27.89 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6089640 114 - 23771897 AUUACCAGAAAACUGUGCAAAGCCUUAACGCCAUAGGUAUGUGUAUAUAUUUAUAUGUUUUGCU------UAUGUGUGCAUGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUG ..............(((((....((..((..((((((((.(..((((....))))..)..))))------)))).((((.((((((((...)))))))).))))..))..)))))))... ( -30.90) >DroSec_CAF1 37999 120 - 1 AUUACCAGAAAACUGUGCAAAGCCUUUACAACAUAGGUAUGUGUAUAUUUUUAUAUGUUUUGCAUAUACAUAUGUGUGCAUGCAUAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUG ..............(((((((..........((((.(((((..(((((...((((((.....))))))...)))))..))))).)))).....)))))))((((((...))))))..... ( -31.36) >DroSim_CAF1 39314 120 - 1 AUUACCAGAAAACUGUGCAAAGCCUUUACAACAUAGGUAUGUGUAUAUUUUUAUAUGUUUUGCAUAUACAUAUGUGUGCAUGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUG ..............(((((....(((.((((((((.(((((((((...............))))))))).)))))((((.((((((((...)))))))).)))).))).))))))))... ( -33.56) >consensus AUUACCAGAAAACUGUGCAAAGCCUUUACAACAUAGGUAUGUGUAUAUUUUUAUAUGUUUUGCAUAUACAUAUGUGUGCAUGCAGAUGCUUCAUUUGCGCGCAUUUGUCGAGUGCACUUG ..............(((((((..........(((..(((((..(((((...((((((.....))))))...)))))..)))))..))).....)))))))((((((...))))))..... (-26.23 = -27.89 + 1.67)


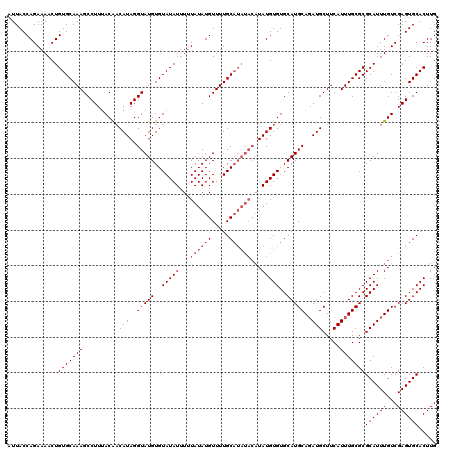
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:51 2006