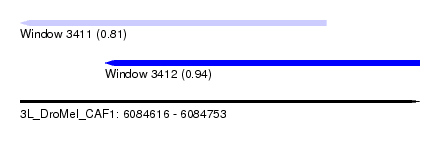
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,084,616 – 6,084,753 |
| Length | 137 |
| Max. P | 0.942791 |

| Location | 6,084,616 – 6,084,721 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.94 |
| Mean single sequence MFE | -15.78 |
| Consensus MFE | -12.86 |
| Energy contribution | -12.72 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
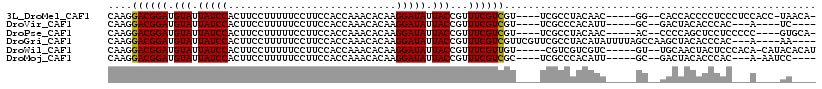
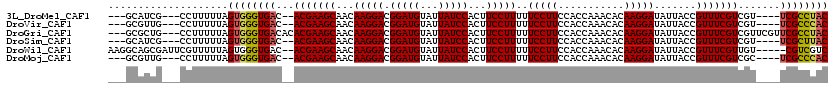
>3L_DroMel_CAF1 6084616 105 - 23771897 CAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU----UCGCCUACAAC-----GG--CACCACCCCUCCCUCCACC-UAACA- ...(((.(((.(((.(((((............................))))).)))......(((((----(.......)))-----))--)........))).)))...-.....- ( -18.59) >DroVir_CAF1 53215 96 - 1 CAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU----UCGCCCACAUU-----GC--GACUACACCCAC---A----UC---- ....((((((.(((.(((((............................))))).)))...))))))((----((((.......-----))--))..))......---.----..---- ( -16.59) >DroPse_CAF1 39063 102 - 1 CAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU----UCGCCUACAAC-----AC--CCCCAGCUCCUCCCCC----GUGCA- .((.((((((.(((.(((((............................))))).)))...)))))).)----)..........-----..--.....((.........----..)).- ( -13.89) >DroGri_CAF1 43664 107 - 1 CAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGUUCGUUCGCCUACAUAUUUUAGCCAAGCUACACCCAC---A----AA---- .((.((((((.(((.(((((............................))))).)))...)))))).)).................((((...)))).......---.----..---- ( -14.49) >DroWil_CAF1 54577 105 - 1 CAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUUGU-----CGUCGUCGUC-----GU--UGCAACUACUCCCACA-CAUACACAU .(((((.(((((...)))))...)))))..(((((...........)))))............(((((-----...((....)-----).--.)))))..........-......... ( -14.50) >DroMoj_CAF1 42793 99 - 1 CAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGC----UCGCCCACAUU-----GC--GACUACACCCAC---A-AAUCC---- .(((((.(((((...)))))...)))))..(((((...........)))))............(((((----...........-----))--))).........---.-.....---- ( -16.60) >consensus CAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU____UCGCCUACAUC_____GC__CACUACACCCAC___A____AC____ ....((((((.(((.(((((............................))))).)))...)))))).................................................... (-12.86 = -12.72 + -0.14)
| Location | 6,084,645 – 6,084,753 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
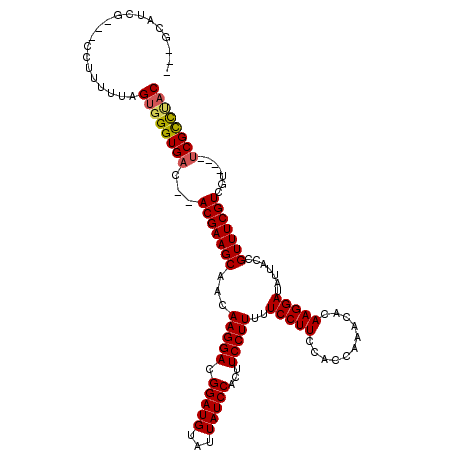
>3L_DroMel_CAF1 6084645 108 - 23771897 ---GCAUCG---CCUUUUUAGUGGGUGAC--ACGAAGCAACAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU----UCGCCUAC ---......---........(((((((((--(((((((...(((((.(((((...)))))...)))))..(((((...........))))).......))))))).).----)))))))) ( -29.50) >DroVir_CAF1 53235 108 - 1 ---GCGUUG---CCUUUUUAGUGGGUGAC--ACGAAGCAACAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU----UCGCCCAC ---......---........(((((((((--(((((((...(((((.(((((...)))))...)))))..(((((...........))))).......))))))).).----)))))))) ( -31.80) >DroGri_CAF1 43691 114 - 1 ---GCGCUG---CCUUUUUAGUGGGUGACACACGAAGCAACAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGUUCGUUCGCCUAC ---......---........((((((((.(((((((((...(((((.(((((...)))))...)))))..(((((...........))))).......))))))).))....)))))))) ( -28.60) >DroSim_CAF1 34327 108 - 1 ---GCAUCG---CCUUUUUAGUGGGUGAC--ACGAAGCAACAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU----UCGCUUAC ---......---........(((((((((--(((((((...(((((.(((((...)))))...)))))..(((((...........))))).......))))))).).----)))))))) ( -26.70) >DroWil_CAF1 54607 113 - 1 AAGGCAGCGAUUCGUUUUUAGUGGGUGAC--ACGAAGCAACAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUUGU-----CGUCGUC ..((((((((..(((.(((.((((....)--..((((.((.(((((.(((((...)))))...))))).)).))))))).))).))..((......)))..)))))))-----)...... ( -31.70) >DroMoj_CAF1 42816 108 - 1 ---GCGUUG---CCUUUUUAGUGGGUGAC--ACGAAGCAACAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGC----UCGCCCAC ---......---........(((((((((--(((((((...(((((.(((((...)))))...)))))..(((((...........))))).......))))))).).----)))))))) ( -31.80) >consensus ___GCAUCG___CCUUUUUAGUGGGUGAC__ACGAAGCAACAAGGACGGAUGUAUUAUCCACUUCCUUUUUCCUUCCACCAAACACAAGGAUAUUACCGUUUCGUCGU____UCGCCUAC ....................((((((((...(((((((...(((((.(((((...)))))...)))))..(((((...........))))).......))))))).......)))))))) (-25.94 = -26.00 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:43 2006