
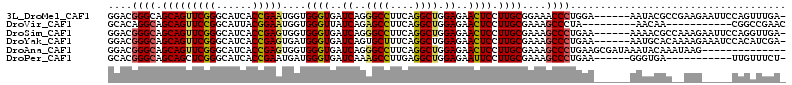
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,999,334 – 5,999,440 |
| Length | 106 |
| Max. P | 0.925786 |

| Location | 5,999,334 – 5,999,440 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.25 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
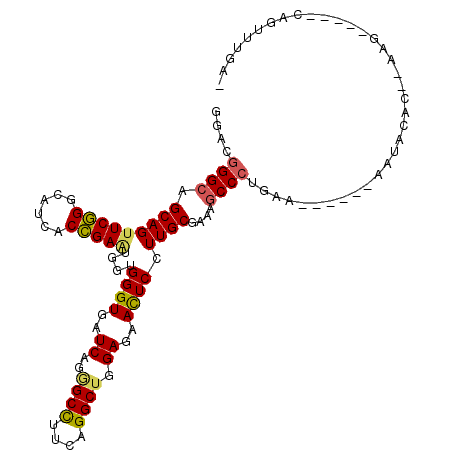
>3L_DroMel_CAF1 5999334 106 + 23771897 -UCAAACUGGAAUUCUUCGGCGUAUU------UCCAGGGUUUCCGCAAGGAGUUCUCCAGCCUGAAGGCCCUGAUCACCCACCAUUCGGUGAUGCCCGAACUGCUGCCCGUCC -....((.((.....(((((.((...------..((((((((((....))).(((........))))))))))((((((........)))))))))))))......)).)).. ( -32.60) >DroVir_CAF1 75885 93 + 1 GUUCGGCCG-----------UUGUU---------UAGGGCUUUCGCAAGGAGUUCUCCAGCCUGAAGGCUCUGAUAACCCACCAUUCCGUAAUGCCGGAACUGCUGCCUGUGC ....(((.(-----------(((((---------.(((((((((((..(((....))).))..)))))))))))))))......(((((......))))).....)))..... ( -29.00) >DroSim_CAF1 58660 106 + 1 -UCAACCUGGAAUUCUUUGGCGUUUU------UUCAGGGCUUUCGCAAGGAGUUCUCCAGCCUGAAGGCCCUGAUCACCCACCACUCGGUGAUGCCCGAACUGCUGCCCGUCC -.......((.....(((((.((...------.(((((((((((((..(((....))).))..)))))))))))(((((........))))).)))))))......))..... ( -33.60) >DroYak_CAF1 58179 106 + 1 -UCGAUGUGGAUUUCUUUUGUGCAUU------UUCAGGGCUUUCGCAAGGAGUUCUCCAGCCUGAAAGCACUGAUCACCCAUCACUCGGUGAUGCCCGAACUGCUGCCCGUCC -..((((.((......((((.(((..------.((((.((((((((..(((....))).))..)))))).))))(((((........)))))))).))))......)))))). ( -32.60) >DroAna_CAF1 54317 99 + 1 --------------CUUAUUUGUAUUUAUCGCUUCAGGGCUUUCGCAAGGAGUUCUCCAGCCUGAAGGCCCUGAUCACCCACCACUCGGUGAUGCCCGAACUGCUGCCCGUCC --------------.......(((.((...((.(((((((((((((..(((....))).))..)))))))))))(((((........))))).))...)).)))......... ( -30.40) >DroPer_CAF1 69415 95 + 1 -AGAAACAA-----------UCACCC------UUCAGGGCUUUCGCAAGGAAUUCUCCAGCCUCAAGGCUUUGAUCACCCAUCAUUCGGUGAUGCCCGAGCUGCUGCCCGUGC -........-----------......------....((((((((....)))).....((((.((..(((....((((((........))))))))).))))))..)))).... ( -27.70) >consensus _UCAAACUG_____CUU__GUGUAUU______UUCAGGGCUUUCGCAAGGAGUUCUCCAGCCUGAAGGCCCUGAUCACCCACCACUCGGUGAUGCCCGAACUGCUGCCCGUCC ....................................((((.(((....)))((((....(((....)))....((((((........))))))....))))....)))).... (-22.77 = -22.88 + 0.11)



| Location | 5,999,334 – 5,999,440 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.25 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -25.09 |
| Energy contribution | -24.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5999334 106 - 23771897 GGACGGGCAGCAGUUCGGGCAUCACCGAAUGGUGGGUGAUCAGGGCCUUCAGGCUGGAGAACUCCUUGCGGAAACCCUGGA------AAUACGCCGAAGAAUUCCAGUUUGA- ((((((((....)))))(((.(((((........)))))((((((..(((..((.(((....)))..)).))).)))))).------.....))).......))).......- ( -36.20) >DroVir_CAF1 75885 93 - 1 GCACAGGCAGCAGUUCCGGCAUUACGGAAUGGUGGGUUAUCAGAGCCUUCAGGCUGGAGAACUCCUUGCGAAAGCCCUA---------AACAA-----------CGGCCGAAC .....(((.((.((((((......)))))).))(((((.((..((((....))))(((....)))....)).)))))..---------.....-----------..))).... ( -32.00) >DroSim_CAF1 58660 106 - 1 GGACGGGCAGCAGUUCGGGCAUCACCGAGUGGUGGGUGAUCAGGGCCUUCAGGCUGGAGAACUCCUUGCGAAAGCCCUGAA------AAAACGCCAAAGAAUUCCAGGUUGA- ((((((((....)))))(((.(((((........)))))(((((((.(((..((.(((....)))..))))).))))))).------.....))).......))).......- ( -39.90) >DroYak_CAF1 58179 106 - 1 GGACGGGCAGCAGUUCGGGCAUCACCGAGUGAUGGGUGAUCAGUGCUUUCAGGCUGGAGAACUCCUUGCGAAAGCCCUGAA------AAUGCACAAAAGAAAUCCACAUCGA- ((((((((....))))).(.((((((.(....).))))))).(((((((((((..(((....)))..((....))))))))------)..))))........))).......- ( -37.10) >DroAna_CAF1 54317 99 - 1 GGACGGGCAGCAGUUCGGGCAUCACCGAGUGGUGGGUGAUCAGGGCCUUCAGGCUGGAGAACUCCUUGCGAAAGCCCUGAAGCGAUAAAUACAAAUAAG-------------- ...(((((....))))).((.(((((........)))))(((((((.(((..((.(((....)))..))))).))))))).))................-------------- ( -33.50) >DroPer_CAF1 69415 95 - 1 GCACGGGCAGCAGCUCGGGCAUCACCGAAUGAUGGGUGAUCAAAGCCUUGAGGCUGGAGAAUUCCUUGCGAAAGCCCUGAA------GGGUGA-----------UUGUUUCU- ((.(((((....))))).)).(((((........)))))(((...((((.(((..((......))..((....))))).))------)).)))-----------........- ( -32.20) >consensus GGACGGGCAGCAGUUCGGGCAUCACCGAAUGGUGGGUGAUCAGGGCCUUCAGGCUGGAGAACUCCUUGCGAAAGCCCUGAA______AAUACAC__AAG_____CAGUUUGA_ ....((((.(((((((((......)))))....((((..((..((((....)))).))..)))).))))....)))).................................... (-25.09 = -24.87 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:27 2006