
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,998,550 – 5,998,650 |
| Length | 100 |
| Max. P | 0.644465 |

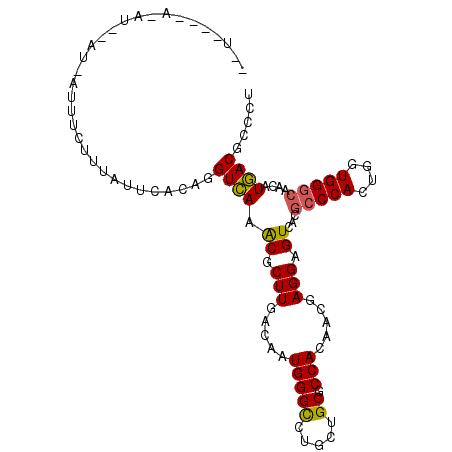
| Location | 5,998,550 – 5,998,650 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -17.25 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5998550 100 + 23771897 -CU----AUAUCUAUGAUUUCUUUCUCUGUAGGUCAAACGCUUGACAAUGGGCUUGCUGCGCCAAAACGAGGAGUCACGCCCAUUGGUGGGCGACAUGACUCCUU -((----(((.....((......))..)))))(((((....)))))..(((((.....)).)))....(((((((((((((((....))))))...))))))))) ( -34.50) >DroPse_CAF1 65782 87 + 1 ----------------AUUUCU--GUUCACAGGUCAAACGCUUAACUAUGGGUCUGCUGCGCCACAACGAGGAGUCCCGCCCACUGGUGGGCAACAUGACGCCCU ----------------....((--(....)))((((.............(((.((.((.((......)))).)).)))(((((....)))))....))))..... ( -27.20) >DroEre_CAF1 54075 105 + 1 CCUAAAGAAAUACAUGAUUUCUUUAUCUGCAGGUCAAACGCUUGACAAUGGGCUUGCUGCGCCAAAACGAGGAGUCACGCCCACUGGUGGGCGACAUGACCCCUU ..(((((((((.....))))))))).......(((((....)))))..(((((.....)).)))....((((.((((((((((....))))))...)))).)))) ( -34.80) >DroYak_CAF1 57406 105 + 1 CCUAAAGAAAUAUAUUAUUUCUUUCUGUGUAGGUCAAGCGCUUGACAAUGGGCUUGCUGCGCCAGAACGAGGAGUCACGCCCACUGGUGGGCGACAUGACCCCCU ....((((((((...))))))))(((((((((..(((((.(........).)))))))))).))))....((.((((((((((....))))))...)))).)).. ( -37.30) >DroMoj_CAF1 59824 102 + 1 --A-AAGCCAUUCUCAACCACUUUAUUCACAGGUUAAGCGCUUGACCAUGGGUCUGUUGCGCCACAAUGAGGAGUCACGCCCACUGGUGGCAAACGUAACGCCCC --.-...........................((((((....))))))..((((..((((((((((...(.((.(.....))).)..)))))....))))))))). ( -25.30) >DroPer_CAF1 68692 87 + 1 ----------------AUUUCU--GUUCACAGGUCAAACGCUUAACUAUGGGUCUGCUGCGCCACAACGAGGAGUCCCGCCCACUGGUGGGCAACAUGACGCCCU ----------------....((--(....)))((((.............(((.((.((.((......)))).)).)))(((((....)))))....))))..... ( -27.20) >consensus __U____A_AU__AU_AUUUCUUUAUUCACAGGUCAAACGCUUGACAAUGGGCCUGCUGCGCCACAACGAGGAGUCACGCCCACUGGUGGGCAACAUGACGCCCU ................................((((.((.(((.....(((((.....)).))).....))).))...(((((....)))))....))))..... (-17.25 = -16.83 + -0.41)

| Location | 5,998,550 – 5,998,650 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.18 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
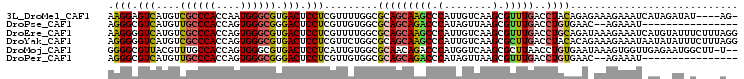
>3L_DroMel_CAF1 5998550 100 - 23771897 AAGGAGUCAUGUCGCCCACCAAUGGGCGUGACUCCUCGUUUUGGCGCAGCAAGCCCAUUGUCAAGCGUUUGACCUACAGAGAAAGAAAUCAUAGAUAU----AG- .((((((((...((((((....)))))))))))))).(((..(((((.((((.....))))...))))).))).........................----..- ( -30.10) >DroPse_CAF1 65782 87 - 1 AGGGCGUCAUGUUGCCCACCAGUGGGCGGGACUCCUCGUUGUGGCGCAGCAGACCCAUAGUUAAGCGUUUGACCUGUGAAC--AGAAAU---------------- ..((.(((.((((((((((.....((((((....)))))))))).)))))))))))...(((((....)))))(((....)--))....---------------- ( -29.60) >DroEre_CAF1 54075 105 - 1 AAGGGGUCAUGUCGCCCACCAGUGGGCGUGACUCCUCGUUUUGGCGCAGCAAGCCCAUUGUCAAGCGUUUGACCUGCAGAUAAAGAAAUCAUGUAUUUCUUUAGG .((((((((...((((((....)))))))))))))).(((..(((((.((((.....))))...))))).))).......(((((((((.....))))))))).. ( -36.10) >DroYak_CAF1 57406 105 - 1 AGGGGGUCAUGUCGCCCACCAGUGGGCGUGACUCCUCGUUCUGGCGCAGCAAGCCCAUUGUCAAGCGCUUGACCUACACAGAAAGAAAUAAUAUAUUUCUUUAGG .((((((((...((((((....)))))))))))))).(((..(((((.((((.....))))...))))).)))........(((((((((...)))))))))... ( -37.90) >DroMoj_CAF1 59824 102 - 1 GGGGCGUUACGUUUGCCACCAGUGGGCGUGACUCCUCAUUGUGGCGCAACAGACCCAUGGUCAAGCGCUUAACCUGUGAAUAAAGUGGUUGAGAAUGGCUU-U-- ((((.((((((((..(.....)..)))))))).))))..(((......)))((((...))))((((.(((((((............)))))))....))))-.-- ( -35.10) >DroPer_CAF1 68692 87 - 1 AGGGCGUCAUGUUGCCCACCAGUGGGCGGGACUCCUCGUUGUGGCGCAGCAGACCCAUAGUUAAGCGUUUGACCUGUGAAC--AGAAAU---------------- ..((.(((.((((((((((.....((((((....)))))))))).)))))))))))...(((((....)))))(((....)--))....---------------- ( -29.60) >consensus AGGGCGUCAUGUCGCCCACCAGUGGGCGUGACUCCUCGUUGUGGCGCAGCAAACCCAUUGUCAAGCGUUUGACCUGCAAACAAAGAAAU_AU__AU_U____A__ .(((.(((....((((((....)))))).))).))).........(((((((((.(........).)))))..))))............................ (-19.40 = -19.18 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:25 2006