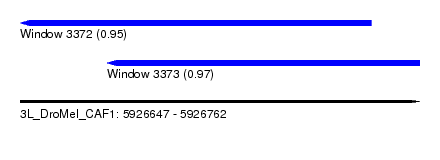
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,926,647 – 5,926,762 |
| Length | 115 |
| Max. P | 0.966296 |

| Location | 5,926,647 – 5,926,748 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.95 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
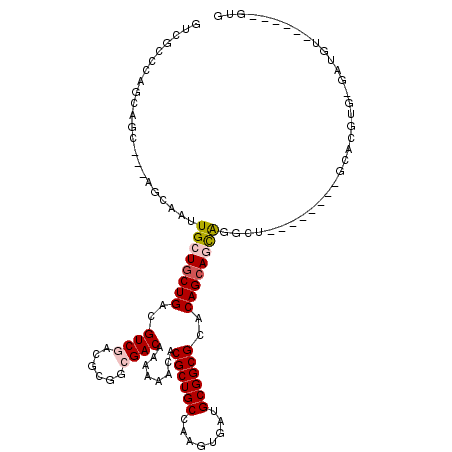
>3L_DroMel_CAF1 5926647 101 - 23771897 GUCGCCCAGCAGC---AGCAAUUGCUGCUGACGUCGACGCGGCGACAAAAAACACGCUGCCAAGUGAUGCGGCGCACAGCAGCAG-CA--------GCAACUG-GCG----ACUUGUG ((((((.((..((---.((...((((((((..((((......))))........((((((........))))))..)))))))))-).--------))..)))-)))----))..... ( -44.20) >DroVir_CAF1 30006 87 - 1 GUCGCCC-------------GCUGCUGCUGGCGUCGACGCGGCGACAAAAAACACGCUGCCAACUGAUGCGGCGCACAGAAAUGAGUU--------GCACGUG-GAUGU--------- (.((((.-------------((....)).)))).)..(((((((((........((((((........))))))..((....)).)))--------)).))))-.....--------- ( -34.10) >DroPse_CAF1 25336 109 - 1 GUCGCCCAGCAGCAACAGCAAUUGCUGCUGACGUCGACGCGGAGACAAAAAACACGCUGCCAAGUGAUGCGGCGCCCAGCAGCAGGCU--------GCACGUG-GAUGUUGCACAGUG ((.((.(((((((((......)))))))))(((((.(((.(....)........((((((........))))))....((((....))--------)).))).-))))).)))).... ( -41.90) >DroEre_CAF1 19037 110 - 1 GUCGCCCAGCAGC---AGCAGUUGCUGCUGACGUCGACGCGGCGACAAAAAACACGCUGCCAAGUGAUGCGGCGCACAGCAGCAG-CAGCACAGCAGCAACUGGGCG----ACUUGUG (((((((((..((---.((.((((((((((.(((((......))))........((((((........))))))....)))))))-))))...)).))..)))))))----))..... ( -55.30) >DroMoj_CAF1 22671 87 - 1 GUCGCCC-------------GCUGCUGCUGGCGUCGACGCGGCGACAAAAAACACGCUGCCAACUGAUGCGGCGCACAGCAACGAGUU--------GCACGUG-GAUGU--------- ((((((.-------------((....))..(((....)))))))))......((((.(((.(((((.(((........))).).))))--------)))))))-.....--------- ( -32.20) >DroPer_CAF1 26838 109 - 1 GUCGCCCAGCAGCAACAGCAAUUGCUGCUGACGUCGACGCGGAGACAAAAAACACGCUGCCAAGUGAUGCGGCGCCCAGCAGCAGGCU--------GCACGUG-GAUGUUGCACAGUG ((.((.(((((((((......)))))))))(((((.(((.(....)........((((((........))))))....((((....))--------)).))).-))))).)))).... ( -41.90) >consensus GUCGCCCAGCAGC___AGCAAUUGCUGCUGACGUCGACGCGGCGACAAAAAACACGCUGCCAAGUGAUGCGGCGCACAGCAGCAGGCU________GCACGUG_GAUGU______GUG ......................((((((((..((((......))))........((((((........))))))..)))))))).................................. (-23.49 = -23.97 + 0.47)

| Location | 5,926,672 – 5,926,762 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -22.38 |
| Energy contribution | -24.88 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5926672 90 - 23771897 GA---------------CAUGUUGCC----------AGU--GUCGCCCAGCAGC---AGCAAUUGCUGCUGACGUCGACGCGGCGACAAAAAACACGCUGCCAAGUGAUGCGGCGCACAG ..---------------..(((((((----------.((--((((.(...((((---(((....)))))))..).))))))))))))).......((((((........))))))..... ( -42.40) >DroVir_CAF1 30027 79 - 1 GA---------------GAUGUUGCC----------A-U--GUCGCCC-------------GCUGCUGCUGGCGUCGACGCGGCGACAAAAAACACGCUGCCAACUGAUGCGGCGCACAG ..---------------..(((((((----------.-(--((((..(-------------(((......)))).))))).))))))).......((((((........))))))..... ( -31.40) >DroEre_CAF1 19071 90 - 1 CA---------------CAUGUUGCC----------AGU--GUCGCCCAGCAGC---AGCAGUUGCUGCUGACGUCGACGCGGCGACAAAAAACACGCUGCCAAGUGAUGCGGCGCACAG ..---------------..(((((((----------.((--((((.(...((((---(((....)))))))..).))))))))))))).......((((((........))))))..... ( -42.40) >DroMoj_CAF1 22692 89 - 1 GA-----GCAACUCGAUGAUGUUGCC----------A-U--GUCGCCC-------------GCUGCUGCUGGCGUCGACGCGGCGACAAAAAACACGCUGCCAACUGAUGCGGCGCACAG (.-----(((((........))))))----------.-(--((((((.-------------((....))..(((....)))))))))).......((((((........))))))..... ( -33.00) >DroAna_CAF1 48587 92 - 1 GA---------------UAUGUUGCU----------AGUGUGUCGCCCAGCAGC---AGCAAUUGCUGCUGACGUCGACGCGGAGACAAAAAACACGCUGCCAAGUGAUGCGGCGCACAG ..---------------..(((.((.----------.((((((((.(...((((---(((....)))))))..).))))..(....).....))))(((((........)))))))))). ( -35.60) >DroPer_CAF1 26868 113 - 1 CACACUGCCA--UA---UAUGUUGCUGCAGCACCAAAAU--GUCGCCCAGCAGCAACAGCAAUUGCUGCUGACGUCGACGCGGAGACAAAAAACACGCUGCCAAGUGAUGCGGCGCCCAG ......(((.--..---..(((..(((((((........--(((((.(((((((((......)))))))))..).))))..(....).........)))))..))..))).)))...... ( -36.70) >consensus GA_______________CAUGUUGCC__________AGU__GUCGCCCAGCAGC___AGCAAUUGCUGCUGACGUCGACGCGGCGACAAAAAACACGCUGCCAAGUGAUGCGGCGCACAG ...................(((.((................(((((((((((((..........))))))).((....)).)))))).........(((((........)))))))))). (-22.38 = -24.88 + 2.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:05 2006