
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,875,949 – 5,876,103 |
| Length | 154 |
| Max. P | 0.734434 |

| Location | 5,875,949 – 5,876,068 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.28 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.00 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
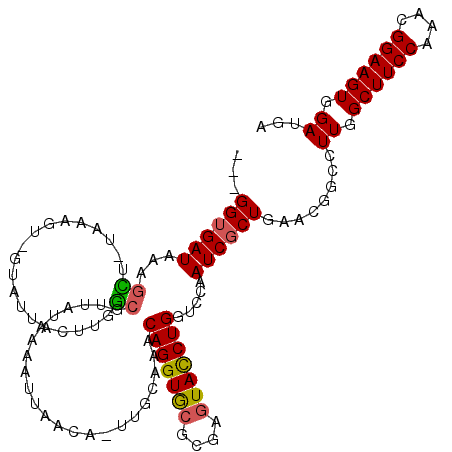
>3L_DroMel_CAF1 5875949 119 - 23771897 AAUAUUAACA-UUUCAAACAGGUGCGCGAGUACCUGGUCCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGAGUUCAAGCAGAGAGUUCCAGUUUUCGGCGAACUCACGUUU ..........-.......(((((((....)))))))((((....(((....)))....((((((....))))))))))((((((..((.(((((......))))).))))))))...... ( -40.60) >DroVir_CAF1 2203 101 - 1 -------------------AGGUGCGCAACUACCUGGUGAAUUCGUUAAAUGGGCUUGGCUUUCAAACGGAAGUGGACGAGUUUAAGCAACGUGUGCCAAUUUUUGGUGAGCUAACGUUU -------------------.((..(((..(((..(..((....))..)..)))(((((((((....((....))....))).))))))...)))..))..........((((....)))) ( -25.80) >DroPse_CAF1 2178 102 - 1 ------------------CAGGUGCGUGACUACCUGGUGCAGUCACUCAAUGGCCUUGGCUUCCAAACGGAAGUCGAUGAGUUCAAGCAGCGUGUGCCAGUUUUAGGCGAACUAACUUUC ------------------..(.(((((((((.(.....).))))))....((..((((((((((....)))))))...)))..)).))).)((.((((.......)))).))........ ( -30.50) >DroGri_CAF1 2225 101 - 1 -------------------AGGUGCGCAAUUACCUGGUCAACUCACUAAAUGGCCUUGGCUUCCACACGGAAGUGGAUGAGUUUAGGCAACGGGUGCCCGUCUUUGGUGAGCUGACGUUU -------------------(((((......))))).((((.(((((((((..((((..((((...(((....)))...))))..)))).((((....)))).))))))))).)))).... ( -43.10) >DroYak_CAF1 2419 119 - 1 AAAAUUAACA-UUGCGAGCAGGUGCGCGAGUACCUGGUGCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGAGUUCAAGCAGAGAGUGCCAGUUUUCGGAGAGCUCACCUUC .........(-(((((..(((((((....))))))).)))))).(((((((..(.((.((((((....)))))).)).).)))).)))...((((.((.......))...))))...... ( -44.70) >DroAna_CAF1 2629 120 - 1 AAAAUUAAAAAAAACCAACAGGUACGCGACUAUCUGGUGCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUAAGUUCAAACAGCGGGUUCCCGUCCUCGGUGAGCUGACGUUC ..................((((((......)))))).......(((((...(..((((((((((....))))))...))))..)...)))))((((((((....))).)))))....... ( -32.10) >consensus __________________CAGGUGCGCGACUACCUGGUGCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGAGUUCAAGCAGCGAGUGCCAGUUUUCGGUGAGCUAACGUUC ..................((((((......))))))((....((((((((.(((.((.((((((....)))))).))...((....))...........)))))))))))....)).... (-20.25 = -20.00 + -0.25)



| Location | 5,875,989 – 5,876,103 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -24.91 |
| Energy contribution | -24.78 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5875989 114 - 23771897 ---GGUGAUAAAGCU-UCAAGU-GUAUUACUUGGCGUUAUAAUAUUAACA-UUUCAAACAGGUGCGCGAGUACCUGGUCCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGA ---..(((....((.-.(((((-(...))))))))((((......)))).-..)))..(((((((....)))))))((((....(((....)))....((((((....)))))))))).. ( -36.20) >DroSec_CAF1 2443 106 - 1 ---GGUGAUAAAGCU-UAAA---------CUUGGCUUUAUAAAAUUAACA-UUGCAAACAGGUGCGCGAGUACCUGGUCCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGA ---....((((((((-....---------...))))))))..........-.......(((((((....)))))))((((....(((....)))....((((((....)))))))))).. ( -36.10) >DroSim_CAF1 2449 114 - 1 ---GGUGAUAAAGCU-UCAAUU-GUAUUUCUUUGCGUUUUAAAAUUAACA-UUGCAAACAGGUGCGCGAGUACCUGGUCCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGA ---(((.....(((.-......-.......((((((..............-.))))))(((((((....)))))))........))).....)))((.((((((....)))))).))... ( -32.66) >DroEre_CAF1 2659 114 - 1 ---GGUGAUAAAGCU-GAAAGU-GUAUUUAUUGGCGUUUUAAAAUCAACA-UUGCGCACAGGUGCGCGAGUACCUGGUACAAUCGCUGAGCGGCCUUGGCUUCCAAACGGAAGUGGAUGA ---(((.....(((.-((...(-((((......((((.............-..)))).(((((((....)))))))))))).))))).....)))((.((((((....)))))).))... ( -37.06) >DroYak_CAF1 2459 115 - 1 ---GGUGAUAAAGUUUUAAACU-GUAUUGCUUGGCAUUAUAAAAUUAACA-UUGCGAGCAGGUGCGCGAGUACCUGGUGCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGA ---((..(((.(((.....)))-.)))..)).(((..............(-(((((..(((((((....))))))).))))))((.....)))))((.((((((....)))))).))... ( -38.10) >DroAna_CAF1 2669 119 - 1 GGGGGGGAUAACAUA-AAAAGUCGUCAAAGCACAAAUGACAAAAUUAAAAAAAACCAACAGGUACGCGACUAUCUGGUGCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUAA ...((.(((..(((.-...((((((((.........)))).............(((....)))....)))).....)))..))).))........((.((((((....)))))).))... ( -23.90) >consensus ___GGUGAUAAAGCU_UAAAGU_GUAUUACUUGGCGUUAUAAAAUUAACA_UUGCAAACAGGUGCGCGAGUACCUGGUCCAAUCGCUGAACGGCCUUGGCUUCCAAACGGAAGUGGAUGA ...((((((...((...................)).......................(((((((....))))))).....))))))........((.((((((....)))))).))... (-24.91 = -24.78 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:52 2006