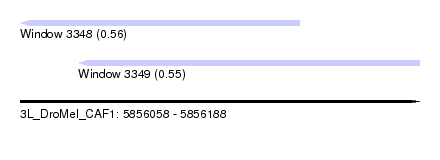
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,856,058 – 5,856,188 |
| Length | 130 |
| Max. P | 0.560929 |

| Location | 5,856,058 – 5,856,149 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -25.03 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5856058 91 - 23771897 AGCAACUCUCCGCA------CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUCCUGC-AGAGAGCAC---CACCCU ...........((.------(((((((.((((((...(((((..(.((((((....)))))).)..))))))))))).))))-.))).))..---...... ( -30.70) >DroPse_CAF1 68647 93 - 1 ----GUUCUCCGCACU----CUCGCGGCACUCGGCCCAUCUGUCCAGAUUCUGCACGGAGUCUGUUCAGAUUCUGCCUCUGCCGGAGAGCACGGACACCCU ----....((((..((----(((.(((((...(((..(((((..((((((((....))))))))..)))))...)))..))))))))))..))))...... ( -44.70) >DroSim_CAF1 46810 91 - 1 AGCAACUCUCCGCA------CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUCCUGC-AGAGAGCAC---CACCCU ...........((.------(((((((.((((((...(((((..(.((((((....)))))).)..))))))))))).))))-.))).))..---...... ( -30.70) >DroEre_CAF1 66728 91 - 1 AGCAACUCUCCGCA------CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUCCUGC-AGAGAGCAC---CACCCU ...........((.------(((((((.((((((...(((((..(.((((((....)))))).)..))))))))))).))))-.))).))..---...... ( -30.70) >DroYak_CAF1 69353 97 - 1 AGCAACUCUCCGCACUCGCACUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUCCUGC-AGAGAGCAC---CACCCU .....(((((.((....))....((((.((((((...(((((..(.((((((....)))))).)..))))))))))).))))-.)))))...---...... ( -32.20) >DroPer_CAF1 67666 93 - 1 ----GUUCUCCGCACU----CUCGCGGCACUCGGCCCAUCUGUCCAGAUUCUGCACGGAGUCUGUUCAGAUUCUGCCUCUGCCGGAGAGCACGGACACCCU ----....((((..((----(((.(((((...(((..(((((..((((((((....))))))))..)))))...)))..))))))))))..))))...... ( -44.70) >consensus AGCAACUCUCCGCA______CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUCCUGC_AGAGAGCAC___CACCCU .....((((((((..........)))(((...((((.(((((..(.((((((....)))))).)..)))))...)))).)))..)))))............ (-25.03 = -24.70 + -0.33)

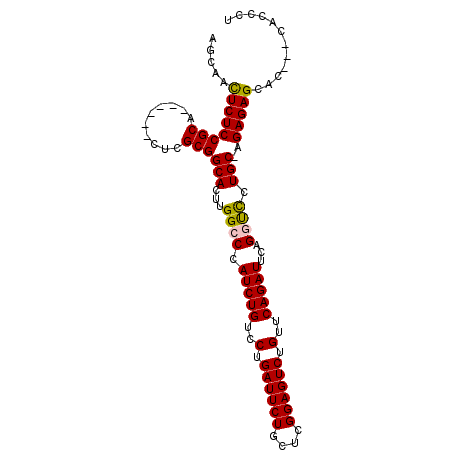
| Location | 5,856,077 – 5,856,188 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
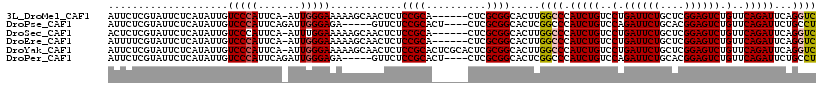
>3L_DroMel_CAF1 5856077 111 - 23771897 AUUCUCGUAUUCUCAUAUUGUCCCAUUCA-AUUGGGAAAAAGCAACUCUCCGCA------CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUC ....................(((((....-..)))))............((((.------...)))).((((((...(((((..(.((((((....)))))).)..))))))))))). ( -29.50) >DroPse_CAF1 68670 109 - 1 AUUCUCGUAUUCUCAUAUUGUCCCAUUCAGAUUGGGAGA-----GUUCUCCGCACU----CUCGCGGCACUCGGCCCAUCUGUCCAGAUUCUGCACGGAGUCUGUUCAGAUUCUGCCU ....................(((((.......)))))((-----((...((((...----...)))).))))(((..(((((..((((((((....))))))))..)))))...))). ( -39.50) >DroSec_CAF1 63268 111 - 1 ACUCUCGUAUUCUCAUAUUGUCCCAUUCA-AUUUGGAAAAAGCAACUCUCCGCA------CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUC .........(((.((.((((.......))-)).)))))...........((((.------...)))).((((((...(((((..(.((((((....)))))).)..))))))))))). ( -25.90) >DroEre_CAF1 66747 111 - 1 AUUUUCGUAUUCUCAUAUUGUCCCAUUCA-AUUGGGAAAAAGCAACUCUCCGCA------CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUC ....................(((((....-..)))))............((((.------...)))).((((((...(((((..(.((((((....)))))).)..))))))))))). ( -29.50) >DroYak_CAF1 69372 117 - 1 AUUCUCGUAUUCUCAUAUUGUCCCAUUCA-AUUGGGAAAAAGCAACUCUCCGCACUCGCACUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUC ....................(((((....-..)))))............((((..........)))).((((((...(((((..(.((((((....)))))).)..))))))))))). ( -28.60) >DroPer_CAF1 67689 109 - 1 AUUCUCGUAUUCUCAUAUUGUCCCAUUCAGAUUGGGAGA-----GUUCUCCGCACU----CUCGCGGCACUCGGCCCAUCUGUCCAGAUUCUGCACGGAGUCUGUUCAGAUUCUGCCU ....................(((((.......)))))((-----((...((((...----...)))).))))(((..(((((..((((((((....))))))))..)))))...))). ( -39.50) >consensus AUUCUCGUAUUCUCAUAUUGUCCCAUUCA_AUUGGGAAAAAGCAACUCUCCGCA______CUCGCGGCACUUGGCCCAUCUGUCCUGAUUCUGCUCGGAGUCUGUUCAGAUUCAGGUC ....................(((((.......)))))............((((..........)))).....((((.(((((..(.((((((....)))))).)..)))))...)))) (-25.04 = -25.10 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:39 2006