
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
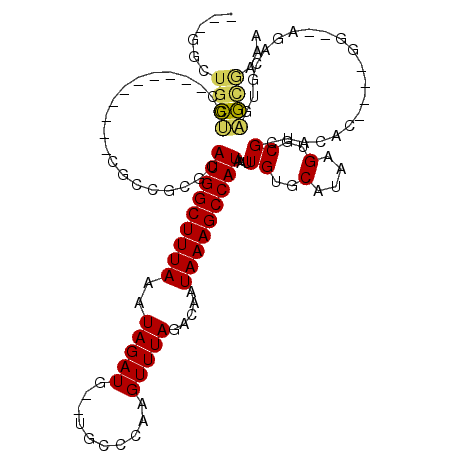
| Location | 5,838,336 – 5,838,438 |
| Length | 102 |
| Max. P | 0.877673 |

| Location | 5,838,336 – 5,838,438 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -14.88 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
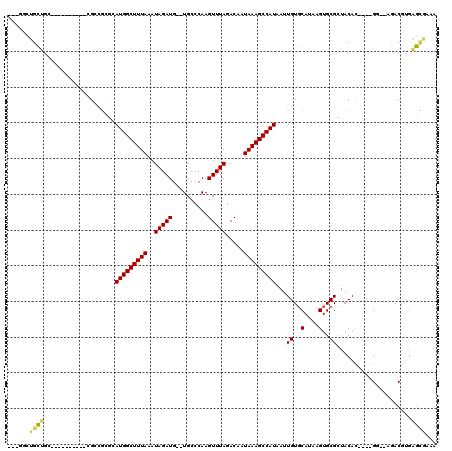
>3L_DroMel_CAF1 5838336 102 + 23771897 GUUGGCUGCUGC----------CGCCGAGCAUGGCUUUAAAUAGAUG--UGCCCAAGUUUAGACAAUAAAGCCAUAAUUGUGCAUAAGUGCGCUACAC----GU--AGACGUGAGCGAAA ...(((....))----------)(((((..(((((((((..(((((.--.......))))).....)))))))))..))).)).......((((.(((----(.--...))))))))... ( -32.60) >DroPse_CAF1 50582 113 + 1 ---GGCUGCUGGAGGAGCCCUCAGAUGCUCAUGGCUUUAAAUAGAUGUGUGCCCAAGUUUAGACAAUAAAGCCAUAAUUGUGCAUAAGUGCGGUACAC----GGUACGACGUGAGCAAAA ---((((.(....).))))......((((((((((((((..(((((.((....)).))))).....)))))))....((((((....(((.....)))----.)))))).)))))))... ( -34.10) >DroGri_CAF1 58564 98 + 1 AG----UGGAGC----------CGCCAGAGAUGGCUUUAAAUAGAUG--UGCCCAAGUUUAGACAAUAAAGCCAUAAUUGUCCAUAAGUGCGGCACAU----GU--ACACAAAACACAAA .(----((....----------.((((....))))......((.(((--((((((..(...((((((.........))))))...)..)).)))))))----.)--)))).......... ( -27.50) >DroYak_CAF1 50709 102 + 1 GUUGGCUGCUGC----------CGCCGCGCAUGGCUUUAAAUAGAUG--UGCCCAAGUUUAGACAAUAAAGCCAUAAUUGUGCAUAAGUGCGCUACAC----GU--AGACGUGAGCGCAA (.((((....))----------)).)(((((((((((((..(((((.--.......))))).....))))))))....))))).....((((((.(((----(.--...)))))))))). ( -37.20) >DroAna_CAF1 49974 100 + 1 ------UGCCGC----------CGCCGAGAAUGGCUUUAAAUAGAUG--UGACCAAGUUUAGACAAUAAAGCCAUAAUUGUGCAUAAGUGCGCUACACCAAGGG--AGACAUGGGCGAGA ------...(((----------((((((..(((((((((..(((((.--.......))))).....)))))))))..))).))....(((.....)))....(.--...)...))))... ( -26.50) >DroPer_CAF1 50765 113 + 1 ---GGCUGCUGGAGGAGCCCUCAGAUGCUCAUGGCUUUAAAUAGAUGUGUGCCCAAGUUUAGACAAUAAAGCCAUAAUUGUGCAUAAGUGCGGUACAC----GGUACGACGUGAGUAAAA ---((((.(....).))))......((((((((((((((..(((((.((....)).))))).....)))))))....((((((....(((.....)))----.)))))).)))))))... ( -32.10) >consensus ___GGCUGCUGC__________CGCCGCGCAUGGCUUUAAAUAGAUG__UGCCCAAGUUUAGACAAUAAAGCCAUAAUUGUGCAUAAGUGCGCUACAC____GG__AGACGUGAGCGAAA ......((((....................(((((((((..(((((..........))))).....)))))))))...((..(....)..)).....................))))... (-14.88 = -14.60 + -0.27)

| Location | 5,838,336 – 5,838,438 |
|---|---|
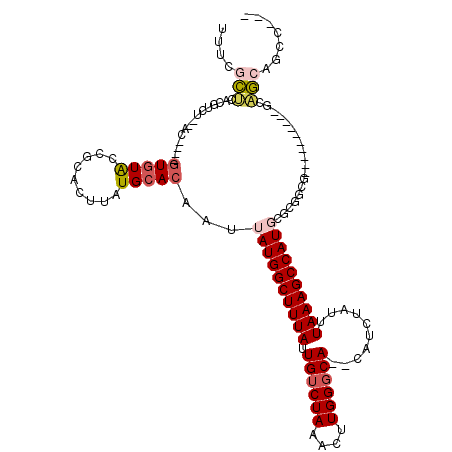
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -15.29 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
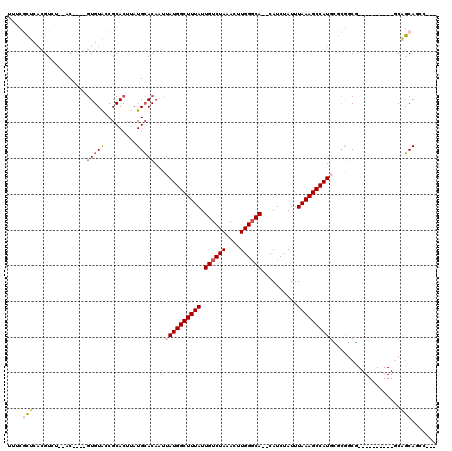
>3L_DroMel_CAF1 5838336 102 - 23771897 UUUCGCUCACGUCU--AC----GUGUAGCGCACUUAUGCACAAUUAUGGCUUUAUUGUCUAAACUUGGGCA--CAUCUAUUUAAAGCCAUGCUCGGCG----------GCAGCAGCCAAC ...((((((((...--.)----))).)))).......((.....((((((((((.((((((....))))))--........))))))))))....))(----------((....)))... ( -31.10) >DroPse_CAF1 50582 113 - 1 UUUUGCUCACGUCGUACC----GUGUACCGCACUUAUGCACAAUUAUGGCUUUAUUGUCUAAACUUGGGCACACAUCUAUUUAAAGCCAUGAGCAUCUGAGGGCUCCUCCAGCAGCC--- ...(((((..((((((..----(((.....))).)))).))....(((((((((.((((((....))))))..........))))))))))))))......((((.(....).))))--- ( -29.10) >DroGri_CAF1 58564 98 - 1 UUUGUGUUUUGUGU--AC----AUGUGCCGCACUUAUGGACAAUUAUGGCUUUAUUGUCUAAACUUGGGCA--CAUCUAUUUAAAGCCAUCUCUGGCG----------GCUCCA----CU ...(((.((((.((--(.----(((((((.((....((((((((.........))))))))....))))))--))).))).))))((((....)))).----------....))----). ( -28.10) >DroYak_CAF1 50709 102 - 1 UUGCGCUCACGUCU--AC----GUGUAGCGCACUUAUGCACAAUUAUGGCUUUAUUGUCUAAACUUGGGCA--CAUCUAUUUAAAGCCAUGCGCGGCG----------GCAGCAGCCAAC .((((((((((...--.)----))).)))))).....((.....((((((((((.((((((....))))))--........))))))))))....))(----------((....)))... ( -36.30) >DroAna_CAF1 49974 100 - 1 UCUCGCCCAUGUCU--CCCUUGGUGUAGCGCACUUAUGCACAAUUAUGGCUUUAUUGUCUAAACUUGGUCA--CAUCUAUUUAAAGCCAUUCUCGGCG----------GCGGCA------ ..(((((.......--.....(((((..((((....))).(((....(((......))).....))))..)--))))........(((......))))----------))))..------ ( -22.90) >DroPer_CAF1 50765 113 - 1 UUUUACUCACGUCGUACC----GUGUACCGCACUUAUGCACAAUUAUGGCUUUAUUGUCUAAACUUGGGCACACAUCUAUUUAAAGCCAUGAGCAUCUGAGGGCUCCUCCAGCAGCC--- .....((((.((.((((.----..)))).))....((((....(((((((((((.((((((....))))))..........))))))))))))))).))))((((.(....).))))--- ( -28.40) >consensus UUUCGCUCACGUCU__AC____GUGUACCGCACUUAUGCACAAUUAUGGCUUUAUUGUCUAAACUUGGGCA__CAUCUAUUUAAAGCCAUGCGCGGCG__________GCAGCAGCC___ ....(((...............(((((.........)))))...((((((((((.((((((....))))))..........))))))))))...................)))....... (-15.29 = -15.98 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:28 2006