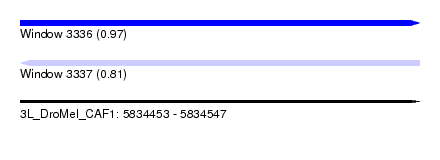
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,834,453 – 5,834,547 |
| Length | 94 |
| Max. P | 0.966327 |

| Location | 5,834,453 – 5,834,547 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
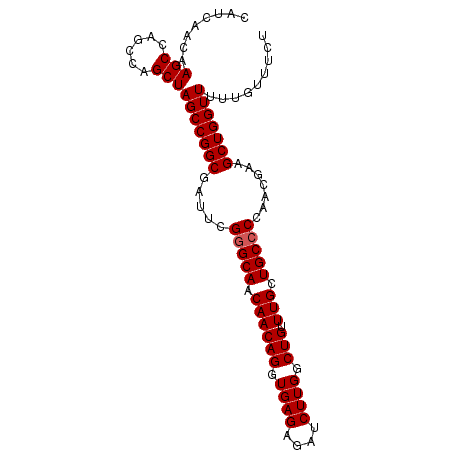
>3L_DroMel_CAF1 5834453 94 + 23771897 CAUCAACAAGCCAGCCAGCUAGCCGGCGAUUCGGGCAACAACAGGUGAGAGAUCUUGGCUGUUUGCUGCCCCAACGAAGCUGGUUUUUGUUUCU ...(((.((((((((..(((....))).....(((((.((((((.((((....)))).))).))).))))).......)))))))))))..... ( -32.50) >DroSec_CAF1 42047 94 + 1 CAUCAACAAGCCAGCCAGCUAGCCGGCGAUUCGGGCAACAACAGGUGAGAGAUCUUGGCUGUUUGCUGCCCCAACGAAGCUGGUUUUUGUUUCU ...(((.((((((((..(((....))).....(((((.((((((.((((....)))).))).))).))))).......)))))))))))..... ( -32.50) >DroSim_CAF1 25050 94 + 1 CAUCAACAAGCCAGCCAGCUAGCCGGCGAUUCGUGCAACAACAGGUGAGAGAUCUUGGCUGUUUGCUGCCCCAACGAAGCUGGUUUUUGUUUCU ...(((.((((((((..(((....)))..((((((((.((((((.((((....)))).))).))).)))....))))))))))))))))..... ( -29.20) >DroEre_CAF1 43166 90 + 1 CAUCAACUAGC----CAGCUAGCCGGCGAUUCGGGCAACAACAGGUGAGAGAUCUUGGCUGUUUGCUGCCCCACCGAAGCUGGUUUUUCUUUCU ........(((----(((((...(((.(....(((((.((((((.((((....)))).))).))).)))))).))).))))))))......... ( -32.20) >DroYak_CAF1 46022 90 + 1 CAUCAACAAGC----CAGCUAGCCGGCGAUUCGGGCAACAACAGGUGAGAGAUCUUGGCUGUUUGCUGCCCCACCGAAGCUGGUUUUUGUUUCU ...(((.((((----(((((...(((.(....(((((.((((((.((((....)))).))).))).)))))).))).))))))))))))..... ( -34.10) >consensus CAUCAACAAGCCAGCCAGCUAGCCGGCGAUUCGGGCAACAACAGGUGAGAGAUCUUGGCUGUUUGCUGCCCCAACGAAGCUGGUUUUUGUUUCU ........(((......)))(((((((.....(((((.((((((.((((....)))).))).))).))))).......)))))))......... (-27.36 = -27.56 + 0.20)


| Location | 5,834,453 – 5,834,547 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -25.88 |
| Energy contribution | -27.28 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
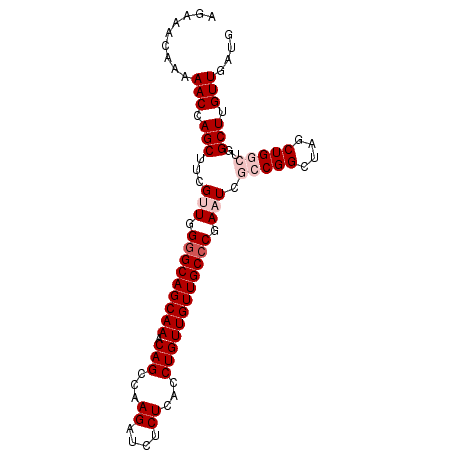
>3L_DroMel_CAF1 5834453 94 - 23771897 AGAAACAAAAACCAGCUUCGUUGGGGCAGCAAACAGCCAAGAUCUCUCACCUGUUGUUGCCCGAAUCGCCGGCUAGCUGGCUGGCUUGUUGAUG ............((((...(((.(((((((((.(((...((....))...)))))))))))).))).(((((((....)))))))..))))... ( -35.20) >DroSec_CAF1 42047 94 - 1 AGAAACAAAAACCAGCUUCGUUGGGGCAGCAAACAGCCAAGAUCUCUCACCUGUUGUUGCCCGAAUCGCCGGCUAGCUGGCUGGCUUGUUGAUG ............((((...(((.(((((((((.(((...((....))...)))))))))))).))).(((((((....)))))))..))))... ( -35.20) >DroSim_CAF1 25050 94 - 1 AGAAACAAAAACCAGCUUCGUUGGGGCAGCAAACAGCCAAGAUCUCUCACCUGUUGUUGCACGAAUCGCCGGCUAGCUGGCUGGCUUGUUGAUG ...(((((...(((((...))))).(((((((.(((...((....))...)))))))))).......(((((((....)))))))))))).... ( -30.50) >DroEre_CAF1 43166 90 - 1 AGAAAGAAAAACCAGCUUCGGUGGGGCAGCAAACAGCCAAGAUCUCUCACCUGUUGUUGCCCGAAUCGCCGGCUAGCUG----GCUAGUUGAUG ...........(((((((((((((((((((((.(((...((....))...))))))))))))....))))))..)))))----).......... ( -34.30) >DroYak_CAF1 46022 90 - 1 AGAAACAAAAACCAGCUUCGGUGGGGCAGCAAACAGCCAAGAUCUCUCACCUGUUGUUGCCCGAAUCGCCGGCUAGCUG----GCUUGUUGAUG ...(((((...(((((((((((((((((((((.(((...((....))...))))))))))))....))))))..)))))----).))))).... ( -36.60) >consensus AGAAACAAAAACCAGCUUCGUUGGGGCAGCAAACAGCCAAGAUCUCUCACCUGUUGUUGCCCGAAUCGCCGGCUAGCUGGCUGGCUUGUUGAUG .........(((.(((...(((.(((((((((.(((...((....))...)))))))))))).))).(((((....)))))..))).))).... (-25.88 = -27.28 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:27 2006