
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
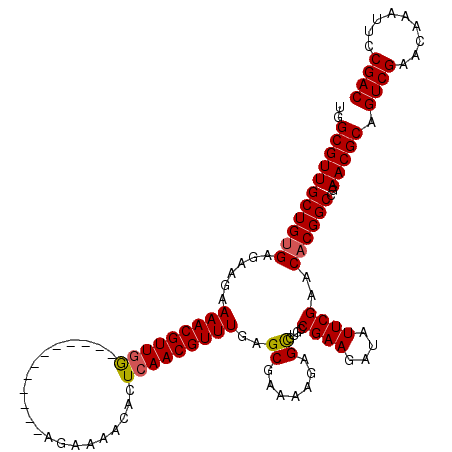
| Location | 5,819,720 – 5,819,828 |
| Length | 108 |
| Max. P | 0.940094 |

| Location | 5,819,720 – 5,819,828 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.47 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.06 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5819720 108 + 23771897 GUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCAUUUCGCUCAAACGUUGAGUGUUUUCU------------CCAACGUUUUCUUCUCACAGCAACGCCA ((((((.....)))))).(((((.(..(((((.((((....)))))))))..).....(((.((((((((((......))------------.)))))))).........)))))))).. ( -29.50) >DroSec_CAF1 27488 108 + 1 GUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCU------------CCAACGUUUUCUUCUCACAGCAACGCCA ((((((.....)))))).(((((.(..(((((.((((....)))))))))..).....(((.((((((((((......))------------.)))))))).........)))))))).. ( -29.00) >DroSim_CAF1 9869 108 + 1 GUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCU------------CCAACGUUUUCUUCUCGCAGCAACGCCA ((((((.....)))))).((((((.(((((((.((((....)))))))))............((((((((((......))------------.)))))))).......)).).))))).. ( -29.80) >DroEre_CAF1 27910 105 + 1 GUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACACUCUUUUCGCUCAAACGUUGAGUGUUUUCU------------UCAACGUUUU---CUCACAGCAACGCCA ((((((.....)))))).......(((.(((((((((....))))).)))).......(((.((((((((((.......)------------))))))))).---.....)))...))). ( -28.70) >DroYak_CAF1 30004 117 + 1 GUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACACUCAUUUCGCUCAAACGUUGAGUGUUUUUUUUUUUUUUUGGUUUAACGUUUU---CUCACAGCAACGCCA ...((...(((((.((..(((((..((((((((((((....))))).))))......((((((.....))))))...............)))..)))))..)---)....)))))..)). ( -29.80) >consensus GUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCU____________CCAACGUUUUCUUCUCACAGCAACGCCA ((((((.....)))))).((((...((.(((((((((....))))).))))......((((((.....)))))).....................................)).)))).. (-25.30 = -25.06 + -0.24)

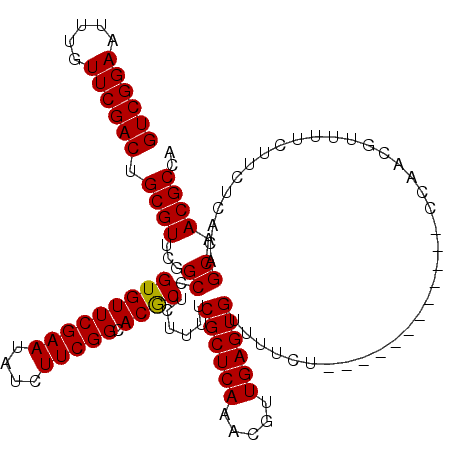

| Location | 5,819,720 – 5,819,828 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.47 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5819720 108 - 23771897 UGGCGUUGCUGUGAGAAGAAAACGUUGG------------AGAAAACACUCAACGUUUGAGCGAAAUGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCAGUCGAACAAAUUCCGAC ..(((((((((((......((((((((.------------((......))))))))))..((.......))....((((....))))..))))))..))))).((((.........)))) ( -30.10) >DroSec_CAF1 27488 108 - 1 UGGCGUUGCUGUGAGAAGAAAACGUUGG------------AGAAAACACUCAACGUUUGAGCGAAAAGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCAGUCGAACAAAUUCCGAC ..(((((((((((......((((((((.------------((......))))))))))..((.......))....((((....))))..))))))..))))).((((.........)))) ( -30.10) >DroSim_CAF1 9869 108 - 1 UGGCGUUGCUGCGAGAAGAAAACGUUGG------------AGAAAACACUCAACGUUUGAGCGAAAAGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCAGUCGAACAAAUUCCGAC ..(((((.(.((.....(.((((((((.------------((......))))))))))...)........((((.((((....))))..)))))).)))))).((((.........)))) ( -27.40) >DroEre_CAF1 27910 105 - 1 UGGCGUUGCUGUGAG---AAAACGUUGA------------AGAAAACACUCAACGUUUGAGCGAAAAGAGUGUGCCGAAGAUAUUCGAACACGGCCGAACGCAGUCGAACAAAUUCCGAC ..(((((((((((.(---.(((((((((------------.........)))))))))...).....((((((.......))))))...))))))..))))).((((.........)))) ( -30.10) >DroYak_CAF1 30004 117 - 1 UGGCGUUGCUGUGAG---AAAACGUUAAACCAAAAAAAAAAAAAAACACUCAACGUUUGAGCGAAAUGAGUGUGCCGAAGAUAUUCGAACACGGCCGAACGCAGUCGAACAAAUUCCGAC ..(((((((((((..---...........................(((((((.(((....)))...)))))))..((((....))))..))))))..))))).((((.........)))) ( -30.50) >consensus UGGCGUUGCUGUGAGAAGAAAACGUUGG____________AGAAAACACUCAACGUUUGAGCGAAAAGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCAGUCGAACAAAUUCCGAC ..(((((((((((......(((((((((.....................)))))))))..((.......))....((((....))))..))))))..))))).((((.........)))) (-25.76 = -25.68 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:08 2006