| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,817,429 – 5,817,566 |
| Length | 137 |
| Max. P | 0.879983 |

| Location | 5,817,429 – 5,817,541 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -28.47 |
| Energy contribution | -27.25 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
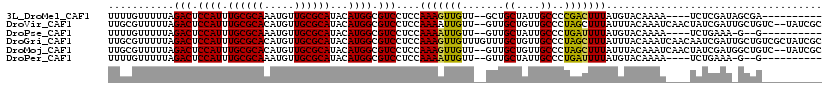
>3L_DroMel_CAF1 5817429 112 + 23771897 UG------AUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUUUUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAGUUGUU--GCUGCUAUUGCCCCGAC ((------(((........)))))..((((.((((.((..(((((......(((.((((.((((((.....))))))...)))).)))...))))).....--)).)))).))))..... ( -31.90) >DroVir_CAF1 28094 112 + 1 ------AGAUUGCUAGCGAUAUCAAUGGCAUUAGUUGCAGUUGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU--GUUGCUGUUGCCCUAGC ------.....((((((((((.(((..(((......((....)).......(((.((((.((((((.....))))))...)))).))).........))).--.))).))))))..)))) ( -34.70) >DroPse_CAF1 27568 118 + 1 UGACGUAGAUAAUUAGCAAUAUCAAUGGCAUUAGUGGCUUUUUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU--GUUGCUAUUGCCCUGAU ....................((((..((((.(((..((..(((((......(((.((((.((((((.....))))))...)))).)))...))))).....--))..))).)))).)))) ( -33.40) >DroGri_CAF1 31836 120 + 1 UGACGUAGAUUGCUAGCGAUAUCAAUGGCAUUAGUUGCGGUUGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUCCUCCAAAGUUGUUUGUUUGCUGUUGCCCUAGC ...........(((((.(....)...((((.((((.((((..((.......(((.((((.((((((.....))))))...)))).))).......))...))))..)))).))))))))) ( -32.84) >DroMoj_CAF1 30163 118 + 1 UGACGUAGAUUGCUAGCGAUAUCAAUGGCAUUAGUUGCGGUUGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUCCUCCAAAGUUGUU--GUUGCUGUUGCCCUAGC ...........((((((((((.(((..(((.(((..(((....)))..)))(((.((((.((((((.....))))))...)))).))).........))).--.))).))))))..)))) ( -35.70) >DroPer_CAF1 27655 118 + 1 UGACGUAGAUAAUUAGCAAUAUCAAUGGCAUUAGUGGCUUUUUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU--GUUGCUAUUGCCCUGAU ....................((((..((((.(((..((..(((((......(((.((((.((((((.....))))))...)))).)))...))))).....--))..))).)))).)))) ( -33.40) >consensus UGACGUAGAUAACUAGCAAUAUCAAUGGCAUUAGUUGCUGUUGCGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU__GUUGCUAUUGCCCUAAC ...........(((((..........((((.(((..((......(((((..(((.((((.((((((.....))))))...)))).)))....)))))......))..))).))))))))) (-28.47 = -27.25 + -1.22)


| Location | 5,817,429 – 5,817,541 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5817429 112 - 23771897 GUCGGGGCAAUAGCAGC--AACAACUUUGGAGGACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAAAAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAU------CA (((((((((.(((..((--..((....))..((((.((((...((((((.....)))))).)))).))))............))..))).)))).)))))............------.. ( -31.80) >DroVir_CAF1 28094 112 - 1 GCUAGGGCAACAGCAAC--AACAAUUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCAACUGCAACUAAUGCCAUUGAUAUCGCUAGCAAUCU------ (((((((((........--............((((.((((...((((((.....)))))).)))).))))......((....))......))))...(....).))))).....------ ( -32.70) >DroPse_CAF1 27568 118 - 1 AUCAGGGCAAUAGCAAC--AACAAUUUUGGAGGACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAAAAAGCCACUAAUGCCAUUGAUAUUGCUAAUUAUCUACGUCA (((((((((.(((....--.....(((((..((((.((((...((((((.....)))))).)))).)))).....)))))......))).)))).))))).................... ( -29.96) >DroGri_CAF1 31836 120 - 1 GCUAGGGCAACAGCAAACAAACAACUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCAACCGCAACUAAUGCCAUUGAUAUCGCUAGCAAUCUACGUCA (((((((((......................((((.((((...((((((.....)))))).)))).))))......((....))......))))...(....).)))))........... ( -32.40) >DroMoj_CAF1 30163 118 - 1 GCUAGGGCAACAGCAAC--AACAACUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCAACCGCAACUAAUGCCAUUGAUAUCGCUAGCAAUCUACGUCA (((((((((........--............((((.((((...((((((.....)))))).)))).))))......((....))......))))...(....).)))))........... ( -32.40) >DroPer_CAF1 27655 118 - 1 AUCAGGGCAAUAGCAAC--AACAAUUUUGGAGGACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAAAAAGCCACUAAUGCCAUUGAUAUUGCUAAUUAUCUACGUCA (((((((((.(((....--.....(((((..((((.((((...((((((.....)))))).)))).)))).....)))))......))).)))).))))).................... ( -29.96) >consensus GCCAGGGCAACAGCAAC__AACAACUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACAAAAAAGCAACUAAUGCCAUUGAUAUCGCUAACAAUCUACGUCA .....((((...((.......((....))..((((.((((...((((((.....)))))).)))).))))............))......)))).......................... (-24.10 = -24.10 + -0.00)

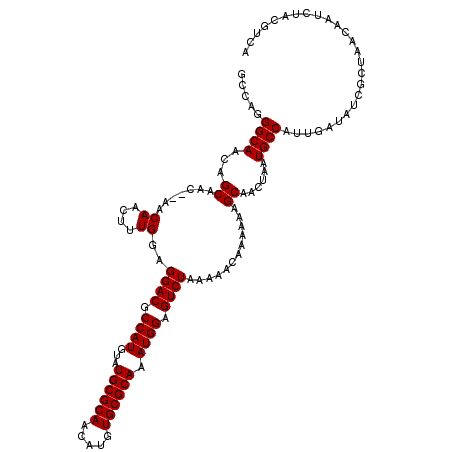

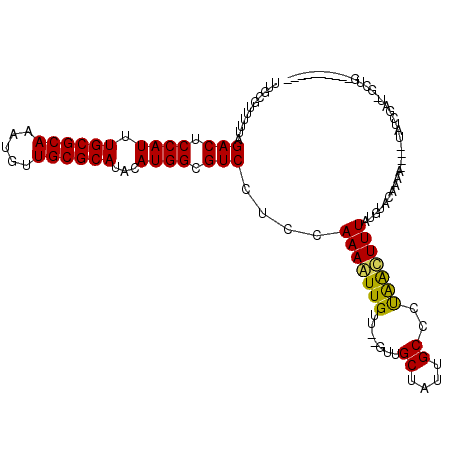
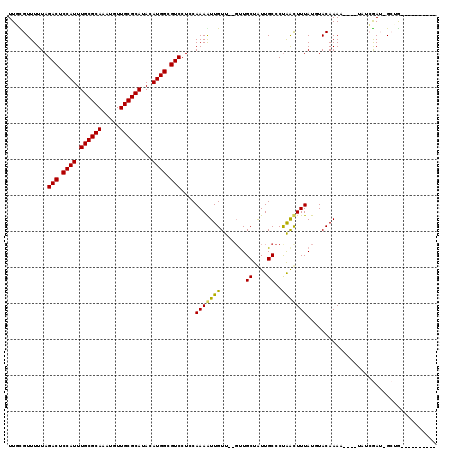
| Location | 5,817,463 – 5,817,566 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -18.67 |
| Energy contribution | -17.87 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5817463 103 + 23771897 UUUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAGUUGUU--GCUGCUAUUGCCCCGACUUUAUGUACAAAA----UCUCGAUAGCGA---------- ((((((.....(((.((((.((((((.....))))))...)))).)))....(((((((..--((.......))..)))))))....))))))----..(((....)))---------- ( -26.30) >DroVir_CAF1 28128 115 + 1 UUGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU--GUUGCUGUUGCCCUAGCUUUAUUUACAAAUCAACUAUCGAUUGCUGUC--UAUCGC ..(((....(((((.((((.((((((.....))))))...))))..........(((((..--((((...(((...............)))..))))...)))))...)))--)).))) ( -24.16) >DroPse_CAF1 27608 100 + 1 UUUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU--GUUGCUAUUGCCCUGAUUUUAUGUACAAAA----UCUGAAA-G--G---------- ...........(((.((((.((((((.....))))))...)))).)))..((.........--...((....))...((((((......))))----)).....-)--)---------- ( -20.40) >DroGri_CAF1 31876 119 + 1 UUGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUCCUCCAAAGUUGUUUGUUUGCUGUUGCCCUAGCUUUAUUUACAAAUCAACAAUCGAUUGCUGUCGCUAUCGC ...........(((.((((.((((((.....))))))...)))).))).......(((((((((..((((......)))).......))))).))))...((((.((....)).)))). ( -30.00) >DroMoj_CAF1 30203 115 + 1 UUGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUCCUCCAAAGUUGUU--GUUGCUGUUGCCCUAGCUUUAUUUACAAAUCAACUAUCGAUGGCUGUC--UAUCGC ..(((....(((((.((((.((((((.....))))))...)))).(((.((...(((((((--((...((..((....))..))...))))..)))))...)).))).)))--)).))) ( -29.70) >DroPer_CAF1 27695 100 + 1 UUUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU--GUUGCUAUUGCCCUGAUUUUAUGUACAAAA----UCUGAAA-G--G---------- ...........(((.((((.((((((.....))))))...)))).)))..((.........--...((....))...((((((......))))----)).....-)--)---------- ( -20.40) >consensus UUGCGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUCCUCCAAAAUUGUU__GUUGCUAUUGCCCUAACUUUAUGUACAAAA____UAUCGAU_GCUG__________ ...........(((.((((.((((((.....))))))...)))).)))....(((((((.......((....))..))))))).................................... (-18.67 = -17.87 + -0.81)
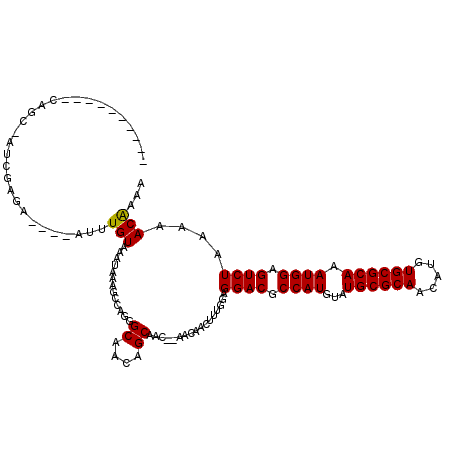
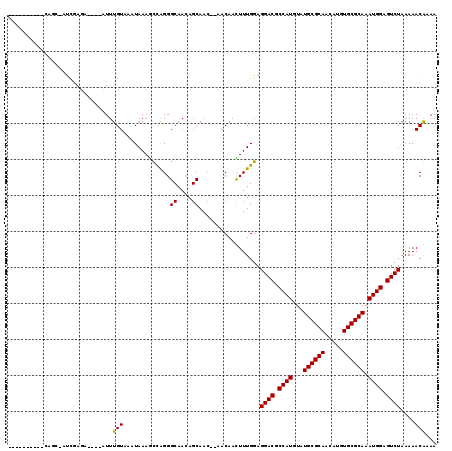
| Location | 5,817,463 – 5,817,566 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -20.35 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5817463 103 - 23771897 ----------UCGCUAUCGAGA----UUUUGUACAUAAAGUCGGGGCAAUAGCAGC--AACAACUUUGGAGGACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAAA ----------..(((((.(.((----(((((....)))))))....).)))))...--......((((..((((.((((...((((((.....)))))).)))).)))).....)))). ( -29.00) >DroVir_CAF1 28128 115 - 1 GCGAUA--GACAGCAAUCGAUAGUUGAUUUGUAAAUAAAGCUAGGGCAACAGCAAC--AACAAUUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCAA ((((((--((((((........)))).))))).......(((..(....))))...--............((((.((((...((((((.....)))))).)))).)))).....))).. ( -30.80) >DroPse_CAF1 27608 100 - 1 ----------C--C-UUUCAGA----UUUUGUACAUAAAAUCAGGGCAAUAGCAAC--AACAAUUUUGGAGGACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAAA ----------(--(-(....((----(((((....))))))))))((....))...--.....(((((..((((.((((...((((((.....)))))).)))).)))).....))))) ( -25.20) >DroGri_CAF1 31876 119 - 1 GCGAUAGCGACAGCAAUCGAUUGUUGAUUUGUAAAUAAAGCUAGGGCAACAGCAAACAAACAACUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCAA (((...((....))..((.(..((((.(((((.......(((..(....))))..)))))))))..).))((((.((((...((((((.....)))))).)))).)))).....))).. ( -34.40) >DroMoj_CAF1 30203 115 - 1 GCGAUA--GACAGCCAUCGAUAGUUGAUUUGUAAAUAAAGCUAGGGCAACAGCAAC--AACAACUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCAA (((.((--(((.(((..(((.(((((.((((....))))(((..(....))))...--..))))))))..)).).((((...((((((.....)))))).)))).)))))....))).. ( -32.60) >DroPer_CAF1 27695 100 - 1 ----------C--C-UUUCAGA----UUUUGUACAUAAAAUCAGGGCAAUAGCAAC--AACAAUUUUGGAGGACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAAA ----------(--(-(....((----(((((....))))))))))((....))...--.....(((((..((((.((((...((((((.....)))))).)))).)))).....))))) ( -25.20) >consensus __________CAGC_AUCGAGA____AUUUGUAAAUAAAGCCAGGGCAACAGCAAC__AACAACUUUGGAGGACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACAAAA .............................(((.............((....)).................((((.((((...((((((.....)))))).)))).))))....)))... (-20.35 = -20.10 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:05 2006