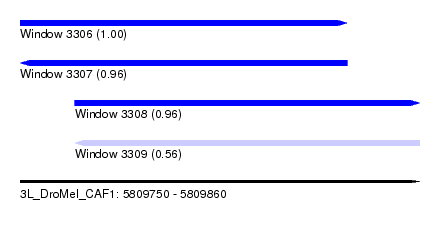
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,809,750 – 5,809,860 |
| Length | 110 |
| Max. P | 0.995988 |

| Location | 5,809,750 – 5,809,840 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -19.08 |
| Energy contribution | -18.95 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5809750 90 + 23771897 GAGCACUUCAAACUGCAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACUGCUGCUUCUGCUUCUGCUUCAUCGUAGCAGA ..(((((.(((.(....).))))))))...((((((((....)))))))).......(((((((....((....))......))))))). ( -32.80) >DroSec_CAF1 17675 84 + 1 AAGCACUUCAAACUGCAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCUAAUAAAACACUGCUGCUUCUGCUUC------UUCGUAGCAGA ..(((((.(((.(....).))))))))...((((((((....)))))))).......(((((((.........------...))))))). ( -28.20) >DroEre_CAF1 17843 77 + 1 AACCACUUCAAACUGCAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACAUCUCU-------UC------UUUAUAGCGCA ..(((..((........)).)))((((....(((((((....)))))))(((((..........-------..------))))).)))). ( -20.40) >DroYak_CAF1 18508 79 + 1 AACCACUUCAAACUGCAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACAUCUCUU-----UUC------UUUAUAGCAGA ............(((((((..(((((....((((((((....)))))))).......)))))...-----.))------).....)))). ( -21.80) >consensus AACCACUUCAAACUGCAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACAGCUCCU_____UUC______UUCAUAGCAGA ............((((((((((....))))))((((((....)))))).....................................)))). (-19.08 = -18.95 + -0.13)


| Location | 5,809,750 – 5,809,840 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -17.99 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5809750 90 - 23771897 UCUGCUACGAUGAAGCAGAAGCAGAAGCAGCAGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUGCAGUUUGAAGUGCUC ((((((.......)))))).((....))...((..(((((((((((((....))))))))((((((.......)))))).)))))..)). ( -30.10) >DroSec_CAF1 17675 84 - 1 UCUGCUACGAA------GAAGCAGAAGCAGCAGUGUUUUAUUAGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUGCAGUUUGAAGUGCUU ((((((.....------..))))))...((((.(.........(((((....)))))(((((((((.......))))))))).).)))). ( -27.20) >DroEre_CAF1 17843 77 - 1 UGCGCUAUAAA------GA-------AGAGAUGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUGCAGUUUGAAGUGGUU ..((((.((((------..-------(((...((((....((((((((....))))))))...))))....)))....)))).))))... ( -23.40) >DroYak_CAF1 18508 79 - 1 UCUGCUAUAAA------GAA-----AAGAGAUGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUGCAGUUUGAAGUGGUU .(..((.((((------...-----.(((...((((....((((((((....))))))))...))))....)))....)))).))..).. ( -23.80) >consensus UCUGCUACAAA______GAA_____AACAGAAGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUGCAGUUUGAAGUGCUU ..((((.(((..............................((((((((....))))))))((((((.......))))))))).))))... (-17.99 = -17.55 + -0.44)


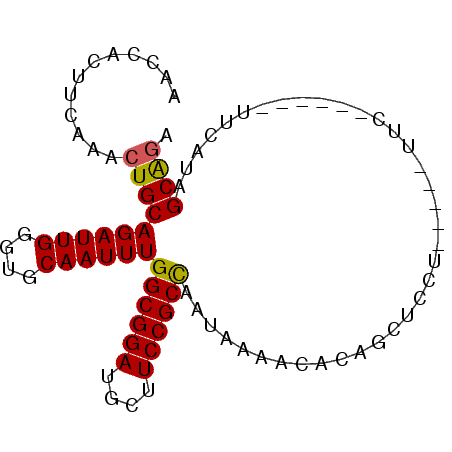
| Location | 5,809,765 – 5,809,860 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -17.25 |
| Energy contribution | -19.50 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5809765 95 + 23771897 CAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACUGCUGCUUCUGCUUCUGCUUCAUCGUAGCAGAGC--AUAUAACA---AUUUUGAGGG .((((((.((((...((((((((....)))))))).......(((((((....((....))......))))))).))--))....))---))))...... ( -33.70) >DroSec_CAF1 17690 89 + 1 CAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCUAAUAAAACACUGCUGCUUCUGCUUC------UUCGUAGCAGAGC--AUACAUCA---AUUUUGAGGG .((((((((((....((((((((....)))))))).....))))..((((.(((((..------.....))))))))--).....))---))))...... ( -28.80) >DroEre_CAF1 17858 84 + 1 CAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACAUCUCU-------UC------UUUAUAGCGCAGCGCAUACAUCA---CUUUUCAGGG .(((..(((((....((((((((....)))))))).......)))))..-------))------).....(((...)))........---.......... ( -21.30) >DroYak_CAF1 18523 88 + 1 CAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACAUCUCUU-----UUC------UUUAUAGCAGAGCAC-UACAUCACUACUUUUCAGGG ..(((..(((((...((((((((....)))))))).........(((((.-----...------.....)).)))))))-)..))).............. ( -23.60) >consensus CAGAUUGGGUGCAAUUUGGCGGAUGCUUCCGCCAAUAAAACACAGCUCCU_____UUC______UUCAUAGCAGAGC__AUACAUCA___AUUUUCAGGG ..(((((.((((...((((((((....)))))))).......(((((((..................))))))).)))).)).))).............. (-17.25 = -19.50 + 2.25)



| Location | 5,809,765 – 5,809,860 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5809765 95 - 23771897 CCCUCAAAAU---UGUUAUAU--GCUCUGCUACGAUGAAGCAGAAGCAGAAGCAGCAGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUG .........(---((((...(--((((((((.......))))).))))..)))))..((((....((((((((....))))))))...))))........ ( -30.90) >DroSec_CAF1 17690 89 - 1 CCCUCAAAAU---UGAUGUAU--GCUCUGCUACGAA------GAAGCAGAAGCAGCAGUGUUUUAUUAGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUG ........((---((.(((.(--((((((((.....------..))))).)))))))((((.......(((((....)))))......)))).))))... ( -24.02) >DroEre_CAF1 17858 84 - 1 CCCUGAAAAG---UGAUGUAUGCGCUGCGCUAUAAA------GA-------AGAGAUGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUG ..((....))---...((((.(((...)))))))..------..-------(((...((((....((((((((....))))))))...))))....))). ( -21.60) >DroYak_CAF1 18523 88 - 1 CCCUGAAAAGUAGUGAUGUA-GUGCUCUGCUAUAAA------GAA-----AAGAGAUGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUG ..((....(((((.(.....-...).)))))....)------)..-----.(((...((((....((((((((....))))))))...))))....))). ( -23.20) >consensus CCCUCAAAAG___UGAUGUAU__GCUCUGCUACAAA______GAA_____AACAGAAGUGUUUUAUUGGCGGAAGCAUCCGCCAAAUUGCACCCAAUCUG .........................................................((((....((((((((....))))))))...))))........ (-15.20 = -15.45 + 0.25)
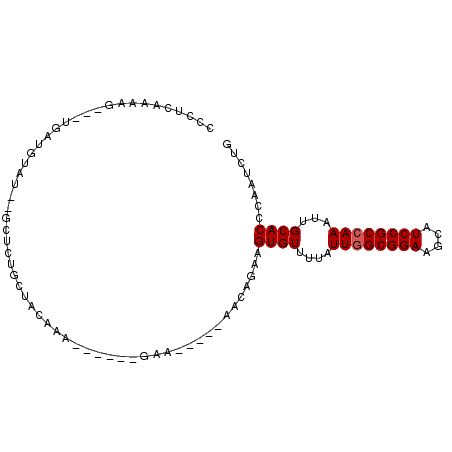
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:00 2006