| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,809,532 – 5,809,663 |
| Length | 131 |
| Max. P | 0.878542 |

| Location | 5,809,532 – 5,809,642 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -28.82 |
| Energy contribution | -29.93 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
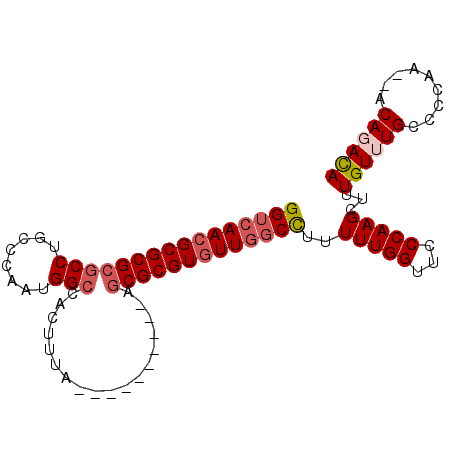
>3L_DroMel_CAF1 5809532 110 + 23771897 AGCACACAUGCAUAUACGCGGACAGAGUAGAGUGUCUGUCUGUGUGGGCCAAACAAAGCUUGGGAACCAAAAAGGCCAACACGCGCUGAAACGUGGUUAAAGUGGCCAUU .(((....)))...((((((((((((........))))))))))))(((((......((((.(....)....)))).(((.((((......)))))))....)))))... ( -36.30) >DroSec_CAF1 17459 110 + 1 AGCACACAUACCAAUACGCGAACAGAGUAGAGUGUCUGUCUGUGUGGGCCAAACAAAGCUUGGGAACCAAAAAGGCCAACACGCGCUGAAACGUGGUUAAAGUGGCCAUU ..((((((.((..((((.(..........).))))..)).))))))(((((......((((.(....)....)))).(((.((((......)))))))....)))))... ( -28.90) >DroEre_CAF1 17638 102 + 1 AGCACACAG----AUACACGGCCAGAGUUGAG----UGUCUGUGUGGGCCAAACAAAGCUUGGGAACCAAAAAGGCCAACACGCGCUGAAACGUGGUUAAAGUGGCCAUU ..(((((((----((((.((((....)))).)----))))))))))(((((......((((.(....)....)))).(((.((((......)))))))....)))))... ( -38.10) >consensus AGCACACAU_C__AUACGCGGACAGAGUAGAGUGUCUGUCUGUGUGGGCCAAACAAAGCUUGGGAACCAAAAAGGCCAACACGCGCUGAAACGUGGUUAAAGUGGCCAUU ..............((((((((((((........))))))))))))(((((......((((.(....)....)))).(((.((((......)))))))....)))))... (-28.82 = -29.93 + 1.11)


| Location | 5,809,568 – 5,809,663 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -26.05 |
| Energy contribution | -26.86 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5809568 95 - 23771897 GGUCAACGCGCGCGCCUGCCCAAUGGCCACUUUAACCACGUUUCAGCGCGUGUUGGCCUUUUUGGUUCCCAAGCUUUGUUUGGCCCAC--ACAGACA ((((((((((((((((........)))......(((...)))...)))))))))))))....(((...((((((...)))))).))).--....... ( -34.50) >DroPse_CAF1 18934 87 - 1 GGUCAACGCGCGCGCCUGCCCAAUGGCCACGUUA----------AGCGCGUGUUGGCCUUUUUGGUUCCCAAGCUUUGUUUGCCACAAGCACAGAUA (((((((((((((((..(((....)))...))..----------.)))))))))))))..(((((...)))))((.((((((...)))))).))... ( -33.20) >DroEre_CAF1 17666 95 - 1 GGUCAACGCGCGCGCCUGCCCAAUGGCCACUUUAACCACGUUUCAGCGCGUGUUGGCCUUUUUGGUUCCCAAGCUUUGUUUGGCCCAC--ACAGACA ((((((((((((((((........)))......(((...)))...)))))))))))))....(((...((((((...)))))).))).--....... ( -34.50) >DroYak_CAF1 18322 95 - 1 GGUCAACGCGCGCGCCUGGCCAAUGGCCACUUUAACCACGUUUCAGCGCGUGUUGGCCUUUUUGGUUCCCAAGCUUUGUUUGGCCCAC--ACACACA (((((((((((((...(((((...)))))....(((...)))...)))))))))))))....(((...((((((...)))))).))).--....... ( -36.80) >DroAna_CAF1 17727 75 - 1 GGUGAAGGCGCGCGCCUGCUCACUGGA-------------------CGCGUGUUG-CUUUUUUGGUUCCCAAGCUUUGUUUGCCCCAA--ACACACA .(((..((((((((((........)).-------------------)))))))).-....(((((....(((((...)))))..))))--))))... ( -22.70) >DroPer_CAF1 18990 87 - 1 GGUCAACGCGCGCGCCUGCCCAAUGGCCACGUUA----------AGCGCGUGUUGGCCUUUUUGGUUCCCAAGCUUUGUUUGCCACAAGCACAGAUA (((((((((((((((..(((....)))...))..----------.)))))))))))))..(((((...)))))((.((((((...)))))).))... ( -33.20) >consensus GGUCAACGCGCGCGCCUGCCCAAUGGCCACUUUA__________AGCGCGUGUUGGCCUUUUUGGUUCCCAAGCUUUGUUUGCCCCAA__ACAGACA ((((((((((((((((........)))..................)))))))))))))..(((((...)))))...((((((.........)))))) (-26.05 = -26.86 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:56 2006