
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,808,756 – 5,808,865 |
| Length | 109 |
| Max. P | 0.999943 |

| Location | 5,808,756 – 5,808,846 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.96 |
| Mean single sequence MFE | -17.42 |
| Consensus MFE | -13.27 |
| Energy contribution | -13.51 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5808756 90 - 23771897 AAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACAAGUGGCCAGGUAGUC-----GC ....((((((........))))))((((....((.(((((.................((.....))(....)..))))).))..))))-----.. ( -17.80) >DroSec_CAF1 16694 90 - 1 AAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACAAGUGGCCAAGUAGUC-----GC ....((((((........))))))((((...(((((((((.................((.....))(....)..)))).)))))))))-----.. ( -18.00) >DroEre_CAF1 16902 83 - 1 AAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAA-------CAGCGGCAACAAGUGGCCAGGUAGUC-----GC ....((((((........))))))((((....((.(((((..((...........-------..))(....)..))))).))..))))-----.. ( -16.42) >DroYak_CAF1 17528 90 - 1 AAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACAAGUGGUCAGGUAGUC-----GC ....((((((........))))))((((...(((((((((.................((.....))(....)..)))).)))))))))-----.. ( -17.70) >DroAna_CAF1 17010 91 - 1 AAAUAUUUUCACACAGACGAAAAUGAUUUCAAGUUGCCACCAGCGGGCAUAAUAACAG----AAGCAGCAACAAGUGGCCAGGUAGUCUGCAGGC .............(((((((((....)))).....(((((..((..((..........----..)).)).....)))))......)))))..... ( -17.20) >consensus AAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACAAGUGGCCAGGUAGUC_____GC ...(((((((........)))))))......(((((((((..((....................))(....)..)))).)))))........... (-13.27 = -13.51 + 0.24)



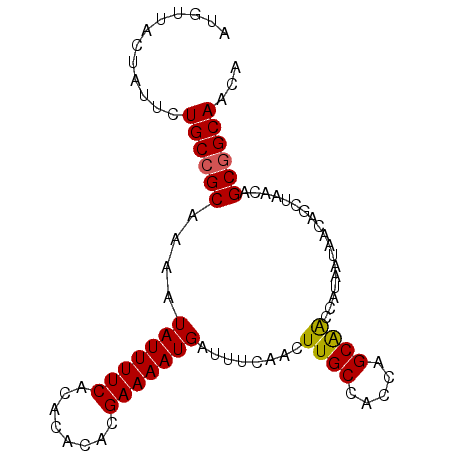
| Location | 5,808,773 – 5,808,865 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5808773 92 + 23771897 UGUUGCCGCUGUUAGCUGUUAUUAUGGUUGCUGGUGGCAAGUUGAAAUCAUUUUCGUGUGUGUGAAAAUAUUUUGCGGCAGAAUAGCAACAU ((((((.((..(((((.............)))))..))..(((((((..(((((((......)))))))..))).))))......)))))). ( -25.52) >DroSec_CAF1 16711 92 + 1 UGUUGCCGCUGUUAGCUGUUAUUAUGGUUGCUGGUGGCAAGUUGAAAUCAUUUUCGUGUGUGUGAAAAUAUUUUGCGGCAGAAUAGCAACAU ((((((.((..(((((.............)))))..))..(((((((..(((((((......)))))))..))).))))......)))))). ( -25.52) >DroEre_CAF1 16919 85 + 1 UGUUGCCGCUG-------UUAUUAUGGUUGCUGGUGGCAAGUUGAAAUCAUUUUCGUGUGUGUGAAAAUAUUUUGCGGCAGAAUAGUAACAA ..(((((((((-------((((((.......))))))))....(((((.(((((((......)))))))))))))))))))........... ( -20.80) >DroYak_CAF1 17545 92 + 1 UGUUGCCGCUGUUAGCUGUUAUUAUGGUUGCUGGUGGCAAGUUGAAAUCAUUUUCGUGUGUGUGAAAAUAUUUUGCGGCAGAAUAGUAACAA ..((((((((..(((((((....)))))))..))))))))(((((((..(((((((......)))))))..))).))))............. ( -24.40) >DroAna_CAF1 17032 88 + 1 UGUUGCUGCUU----CUGUUAUUAUGCCCGCUGGUGGCAACUUGAAAUCAUUUUCGUCUGUGUGAAAAUAUUUUGCGGCAUCUCGCUACCAU ...((((((..----.........(((((....).))))....(((((.(((((((......))))))))))))))))))............ ( -18.90) >consensus UGUUGCCGCUGUUAGCUGUUAUUAUGGUUGCUGGUGGCAAGUUGAAAUCAUUUUCGUGUGUGUGAAAAUAUUUUGCGGCAGAAUAGUAACAU ..(((((((.......((((((((.......))))))))....(((((.(((((((......)))))))))))))))))))........... (-18.50 = -18.94 + 0.44)

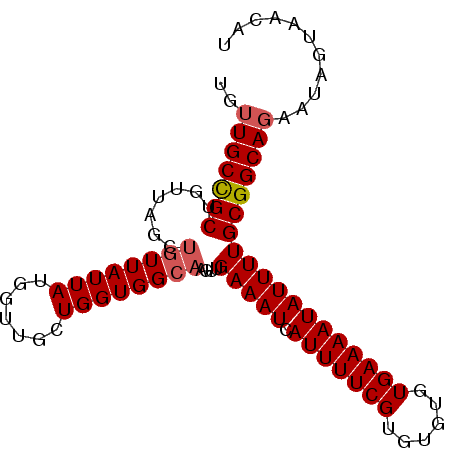

| Location | 5,808,773 – 5,808,865 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -17.46 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.73 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5808773 92 - 23771897 AUGUUGCUAUUCUGCCGCAAAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACA ............((((((....(((((((........)))))))........((((.....))))..................))))))... ( -17.90) >DroSec_CAF1 16711 92 - 1 AUGUUGCUAUUCUGCCGCAAAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACA ............((((((....(((((((........)))))))........((((.....))))..................))))))... ( -17.90) >DroEre_CAF1 16919 85 - 1 UUGUUACUAUUCUGCCGCAAAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAA-------CAGCGGCAACA ............((((((....(((((((........)))))))........((((.....)))).........-------..))))))... ( -17.90) >DroYak_CAF1 17545 92 - 1 UUGUUACUAUUCUGCCGCAAAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACA ............((((((....(((((((........)))))))........((((.....))))..................))))))... ( -17.90) >DroAna_CAF1 17032 88 - 1 AUGGUAGCGAGAUGCCGCAAAAUAUUUUCACACAGACGAAAAUGAUUUCAAGUUGCCACCAGCGGGCAUAAUAACAG----AAGCAGCAACA .((((((((......)))....(((((((........))))))).........)))))...((..((..........----..)).)).... ( -15.70) >consensus AUGUUACUAUUCUGCCGCAAAAUAUUUUCACACACACGAAAAUGAUUUCAACUUGCCACCAGCAACCAUAAUAACAGCUAACAGCGGCAACA ............((((((....(((((((........)))))))........((((.....))))..................))))))... (-16.78 = -16.66 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:53 2006