
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,786,843 – 5,786,950 |
| Length | 107 |
| Max. P | 0.671609 |

| Location | 5,786,843 – 5,786,950 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -23.14 |
| Energy contribution | -22.83 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
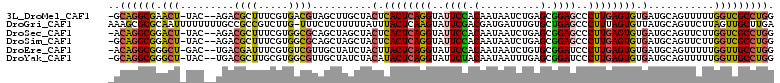
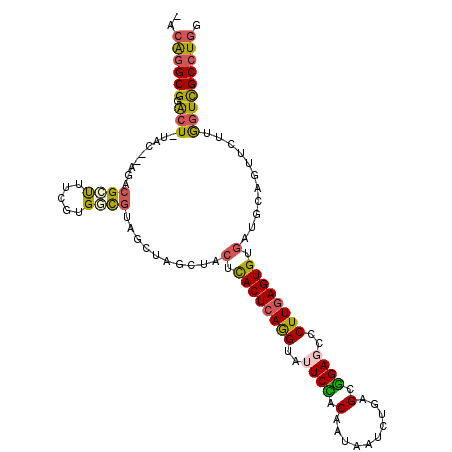
>3L_DroMel_CAF1 5786843 107 + 23771897 -GCAGGCGAACU-UAC--AGACGCUUUCGUGACGUAGCUUGCUACUCACUCAGGUAUUCCACAAUAAUCUGAGCGGAGCCCUUGAGUGUGAUGCAGUUUUUGGUCGCCUGG -.(((((((...-..(--(((.(((..(((.((((((....))))..(((((((..((((.(..........).))))..))))))))).))).))).)))).))))))). ( -38.10) >DroGri_CAF1 191 110 + 1 AAAGCGCGCAAUUUUUUUUGCCGCCGUCUUG-UUUCUCUUUUUAUUUACUCAAGUAUUCGACGAUGAUUUGUGCUGAGCCCUUUAGUGUUAUGCAGUUCUUAGUUGCUUGA ...(((.((((......))))))).......-.................(((((((....((((....))))((((((..((.((......)).))..))))))))))))) ( -21.60) >DroSec_CAF1 183 107 + 1 -ACAGGCGGACU-UAC--AGACGCUUUCGUGGCGCAGCUAGCUACUCACUCAGGUAUUCCACAAUAAUCUGAGCGGAGCCCUUGAGUGUGAUGCAGUUCUUGGUCGCCUGG -.((((((.(((-...--.(((((..(((((((.......))))).((((((((..((((.(..........).))))..)))))))).)).)).)))...))))))))). ( -39.50) >DroSim_CAF1 208 107 + 1 -GCAGGCGGACU-UAC--AGACGCUUUCGUGGCGCAGCUAGCUACUCACUCAGGUAUUCCACAAUAAUCUGAGCGGAGCCCUUGAGUGUGAUGCAGUUCUUGGUCGCCUGG -.((((((.(((-...--.(((((..(((((((.......))))).((((((((..((((.(..........).))))..)))))))).)).)).)))...))))))))). ( -39.20) >DroEre_CAF1 208 107 + 1 -ACAGGCGGGCU-GAC--UGACGAUUUCGUGUCGUUGCUAUCUACUUACUCAGGUAUUCCACAAUAAUCUGUGCGGAUCCCUUGAGUGUGAUGCAGUUUUUGGUUGCCUGG -.(((((((.(.-(((--(((((((.....))))))((.(((....((((((((.((((((((......)))).)))).)).)))))).)))))))))...).))))))). ( -39.90) >DroYak_CAF1 256 107 + 1 -GCAGGCGGGCU-UAC--UGACGCUUGCGUGGCGUUGCUAUCUACAUACUCAGGUAUUCUACAAUAAUUUGAGCGGAUCCCUUGAGUGUGAUGCAGUUUUUGGUUGCCUGG -.(((((((.(.-.((--(((((((.....))))).........((((((((((.((((..(((....)))...)))).)).))))))))...))))....).))))))). ( -36.90) >consensus _ACAGGCGGACU_UAC__AGACGCUUUCGUGGCGUAGCUAGCUACUCACUCAGGUAUUCCACAAUAAUCUGAGCGGAGCCCUUGAGUGUGAUGCAGUUCUUGGUCGCCUGG ..((((((.(((.........((((.....))))..........(.((((((((..((((.(..........).))))..)))))))).)...........))))))))). (-23.14 = -22.83 + -0.30)
| Location | 5,786,843 – 5,786,950 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -19.01 |
| Energy contribution | -20.40 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5786843 107 - 23771897 CCAGGCGACCAAAAACUGCAUCACACUCAAGGGCUCCGCUCAGAUUAUUGUGGAAUACCUGAGUGAGUAGCAAGCUACGUCACGAAAGCGUCU--GUA-AGUUCGCCUGC- .(((((((......((((((...((((((.((..(((((..........)))))...)))))))).((((....))))(.(.(....).).))--)).-)))))))))).- ( -36.10) >DroGri_CAF1 191 110 - 1 UCAAGCAACUAAGAACUGCAUAACACUAAAGGGCUCAGCACAAAUCAUCGUCGAAUACUUGAGUAAAUAAAAAGAGAAA-CAAGACGGCGGCAAAAAAAAUUGCGCGCUUU ..((((.....((..((............))..))..((.........(((((....((((..................-)))).)))))((((......)))))))))). ( -16.07) >DroSec_CAF1 183 107 - 1 CCAGGCGACCAAGAACUGCAUCACACUCAAGGGCUCCGCUCAGAUUAUUGUGGAAUACCUGAGUGAGUAGCUAGCUGCGCCACGAAAGCGUCU--GUA-AGUCCGCCUGU- .((((((((..(((...((....((((((.((..(((((..........)))))...)))))))).((((....)))).........)).)))--...-.)).)))))).- ( -36.90) >DroSim_CAF1 208 107 - 1 CCAGGCGACCAAGAACUGCAUCACACUCAAGGGCUCCGCUCAGAUUAUUGUGGAAUACCUGAGUGAGUAGCUAGCUGCGCCACGAAAGCGUCU--GUA-AGUCCGCCUGC- .((((((((..(((...((....((((((.((..(((((..........)))))...)))))))).((((....)))).........)).)))--...-.)).)))))).- ( -36.70) >DroEre_CAF1 208 107 - 1 CCAGGCAACCAAAAACUGCAUCACACUCAAGGGAUCCGCACAGAUUAUUGUGGAAUACCUGAGUAAGUAGAUAGCAACGACACGAAAUCGUCA--GUC-AGCCCGCCUGU- .(((((..........(((((((((((((.((..((((((........))))))...)))))))..)).))).)))..((((((....)))..--)))-.....))))).- ( -33.30) >DroYak_CAF1 256 107 - 1 CCAGGCAACCAAAAACUGCAUCACACUCAAGGGAUCCGCUCAAAUUAUUGUAGAAUACCUGAGUAUGUAGAUAGCAACGCCACGCAAGCGUCA--GUA-AGCCCGCCUGC- .(((((.........((((((...(((((.((..((..(..........)..))...)))))))))))))...((.((((.......))))..--)).-.....))))).- ( -26.70) >consensus CCAGGCAACCAAAAACUGCAUCACACUCAAGGGCUCCGCUCAGAUUAUUGUGGAAUACCUGAGUAAGUAGAUAGCUACGCCACGAAAGCGUCA__GUA_AGUCCGCCUGC_ .(((((.........((((.....(((((.((..(((((..........)))))...)))))))..))))......((((.......)))).............))))).. (-19.01 = -20.40 + 1.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:38 2006