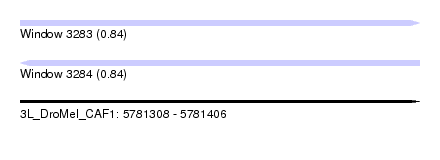
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,781,308 – 5,781,406 |
| Length | 98 |
| Max. P | 0.844983 |

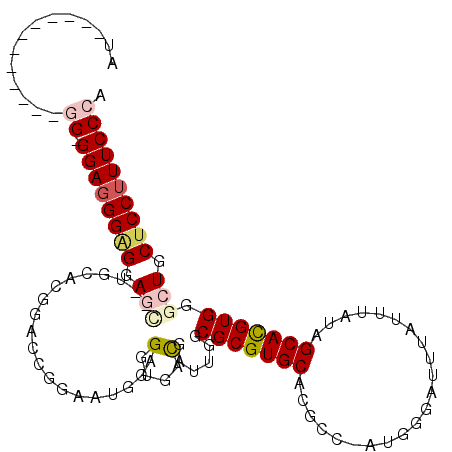
| Location | 5,781,308 – 5,781,406 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -16.99 |
| Energy contribution | -18.55 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
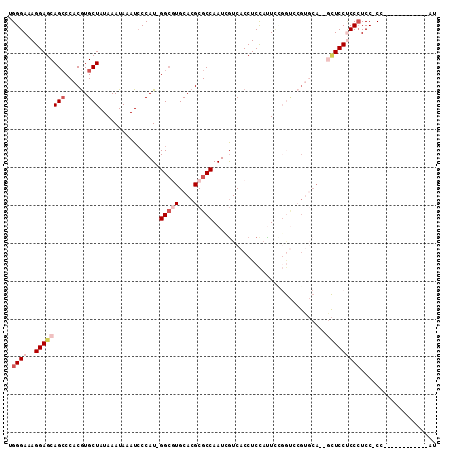
>3L_DroMel_CAF1 5781308 98 + 23771897 UGGGAAAGGAGCAGCCCACGUGCUAUAAAUAAAUCCCAU-GGCGUGCACGCGCCAAUUGUCACCUCCAUUCCGGUCCGUGCA--GCUCCUCCCUCCCCC------------AU .((((.(((((((((......)))..............(-(((((....))))))..((.(((..((.....))...)))))--))))))...))))..------------.. ( -31.30) >DroPse_CAF1 12932 86 + 1 UGGGAAAGGAGCAGCCCACGUGCUAUAAAUAAAUCCCAUCGGGCUGCACGCGCCAAUCGUCACCUCCACUUA--------------UCCCCCCUCC-CU------------AA .((((..((.(((((((..(((..((......))..))).)))))))..(((.....)))............--------------...))..)))-).------------.. ( -22.80) >DroSec_CAF1 6921 97 + 1 UGGGAAAGGAGCAGACCACGUGCUAUAAAUAAAUCCCAU-GGCGUGCACGCGCCAAUCGUCACCUCCAUUCAGGUCCGUGCA--GCUCCUCCCUCC-CC------------AU .((((.((((((....((((..................(-(((((....))))))......((((......)))).))))..--))))))...)))-).------------.. ( -31.10) >DroSim_CAF1 7381 97 + 1 UGGGAAAGGAGCAGCCCACGUGCUAUAAAUAAAUCCCAU-GGCGUGCACGCGCCAAUCGUCACCUCCAUUCCGGUCCGUGCA--GCUCCUCCCUCC-CC------------AU .((((.((((((.........(((((...........))-))).((((((.(((..................))).))))))--))))))...)))-).------------.. ( -31.07) >DroEre_CAF1 13485 109 + 1 UGGGAAAGGAGCAGCCCACGUGCUAUAAAUAAAUCCCAU-GGCGUGCACGCGCCAAUCAUCACCUCCAUUCUGGUCCGAGCA--GCUCCUCCCUCC-CCCUCCGCGUCCCCUU .((((.((((((.((.(.....................(-(((((....))))))......(((........)))..).)).--))))))...)))-)............... ( -30.10) >DroAna_CAF1 12649 106 + 1 UGGGAAAGGAGCAGCCCACAUGCUAUAAAUAAAUGCCAU-GGCCUGCACGCGCCAAUCGUCACCUCUCCUACCACUAGUCCACCAUUCCACCAUCC-CACC---AGUCU--AC (((((.(((.((((.(((...((...........))..)-)).))))..(((.....)))..))).))))).........................-....---.....--.. ( -19.50) >consensus UGGGAAAGGAGCAGCCCACGUGCUAUAAAUAAAUCCCAU_GGCGUGCACGCGCCAAUCGUCACCUCCAUUCCGGUCCGUGCA__GCUCCUCCCUCC_CC____________AU .((((..((((((((......)))................(((((....)))))..............................))))))))).................... (-16.99 = -18.55 + 1.56)

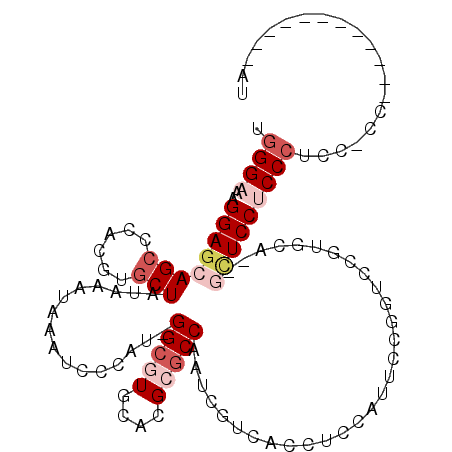
| Location | 5,781,308 – 5,781,406 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -23.25 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5781308 98 - 23771897 AU------------GGGGGAGGGAGGAGC--UGCACGGACCGGAAUGGAGGUGACAAUUGGCGCGUGCACGCC-AUGGGAUUUAUUUAUAGCACGUGGGCUGCUCCUUUCCCA ..------------.((((((((((.(((--(.((((..(..((((((((....)...(((((......))))-).....)))))))...)..)))))))).)))))))))). ( -44.50) >DroPse_CAF1 12932 86 - 1 UU------------AG-GGAGGGGGGA--------------UAAGUGGAGGUGACGAUUGGCGCGUGCAGCCCGAUGGGAUUUAUUUAUAGCACGUGGGCUGCUCCUUUCCCA ..------------.(-(((((((((.--------------..(((...(....)......(((((((..(((...)))((......)).))))))).))).)))))))))). ( -34.20) >DroSec_CAF1 6921 97 - 1 AU------------GG-GGAGGGAGGAGC--UGCACGGACCUGAAUGGAGGUGACGAUUGGCGCGUGCACGCC-AUGGGAUUUAUUUAUAGCACGUGGUCUGCUCCUUUCCCA .(------------((-((((((((.((.--..(((((...(((((((((....)...(((((......))))-).....))))))))...).))))..)).))))))))))) ( -39.20) >DroSim_CAF1 7381 97 - 1 AU------------GG-GGAGGGAGGAGC--UGCACGGACCGGAAUGGAGGUGACGAUUGGCGCGUGCACGCC-AUGGGAUUUAUUUAUAGCACGUGGGCUGCUCCUUUCCCA .(------------((-((((((((.(((--(.((((..(..((((((((....)...(((((......))))-).....)))))))...)..)))))))).))))))))))) ( -44.70) >DroEre_CAF1 13485 109 - 1 AAGGGGACGCGGAGGG-GGAGGGAGGAGC--UGCUCGGACCAGAAUGGAGGUGAUGAUUGGCGCGUGCACGCC-AUGGGAUUUAUUUAUAGCACGUGGGCUGCUCCUUUCCCA ..(....)......((-((((((((.(((--(..(((.(((........)))..)))....(((((((.....-................))))))))))).)))))))))). ( -40.70) >DroAna_CAF1 12649 106 - 1 GU--AGACU---GGUG-GGAUGGUGGAAUGGUGGACUAGUGGUAGGAGAGGUGACGAUUGGCGCGUGCAGGCC-AUGGCAUUUAUUUAUAGCAUGUGGGCUGCUCCUUUCCCA ..--.....---..((-(((....(((...........(((.(((....(....)..))).)))..((((.((-(((((...........)).))))).)))))))..))))) ( -30.70) >consensus AU____________GG_GGAGGGAGGAGC__UGCACGGACCGGAAUGGAGGUGACGAUUGGCGCGUGCACGCC_AUGGGAUUUAUUUAUAGCACGUGGGCUGCUCCUUUCCCA ..............((.((((((((.(((....................(....)......(((((((......................))))))).))).)))))))))). (-23.25 = -24.12 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:36 2006