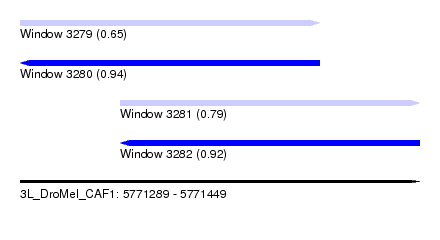
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,771,289 – 5,771,449 |
| Length | 160 |
| Max. P | 0.941762 |

| Location | 5,771,289 – 5,771,409 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -28.60 |
| Energy contribution | -29.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5771289 120 + 23771897 AAACUUAAAAGGCAUGUGCUAAUAAAAGGAAAUUUAUUAAUUAGUUACCGCCUUCAUAGGCUCUCAAACUUCCUCGGCACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUCGGUUGACGG ........(((((....((((((................))))))....)))))....((((((((((((.(....)...)))))))))))).(((.(.((((........))))).))) ( -31.29) >DroSec_CAF1 13888 120 + 1 AAACUUAAAAGGUAUGAGUUAAUAAAGGGAAAUUUAUUAAUUAGUUCCCGUCUUCAUAGGCUCUCAAACCUCCUCGACACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUUGGUUGACGG ..........((....((((((((((......))))))))))....))((((.(((..(((((((((((............)))))))))))..)))..((((........)))))))). ( -34.60) >DroSim_CAF1 13751 120 + 1 AAACUUAAAAGGUAUGAGUUAAUAAAGGGAAAUUUAUUAAUUAGUUCCCGUCUUUAUUGGCUCUCAAACCUCCUCGACACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUUGCUUGACGG ..........((....((((((((((......))))))))))....))((((.(((.((((((((((((............)))))))))))).))).((((.........)))))))). ( -33.30) >consensus AAACUUAAAAGGUAUGAGUUAAUAAAGGGAAAUUUAUUAAUUAGUUCCCGUCUUCAUAGGCUCUCAAACCUCCUCGACACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUUGGUUGACGG ................((((((((((......)))))))))).....(((((......(((((((((((............)))))))))))(((....))).............))))) (-28.60 = -29.60 + 1.00)

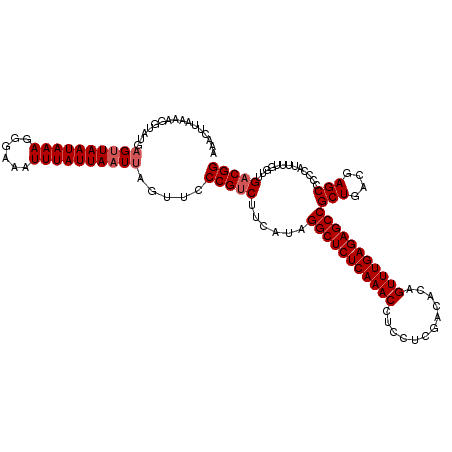

| Location | 5,771,289 – 5,771,409 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -34.11 |
| Energy contribution | -33.23 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5771289 120 - 23771897 CCGUCAACCGAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGCCGAGGAAGUUUGAGAGCCUAUGAAGGCGGUAACUAAUUAAUAAAUUUCCUUUUAUUAGCACAUGCCUUUUAAGUUU (((((..((......))......(((..((((((((((((...(....).))))))))))))..))).))))).((((...(((((((......)))))))((....))......)))). ( -36.20) >DroSec_CAF1 13888 120 - 1 CCGUCAACCAAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGUCGAGGAGGUUUGAGAGCCUAUGAAGACGGGAACUAAUUAAUAAAUUUCCCUUUAUUAACUCAUACCUUUUAAGUUU (((((..((.....)).......(((..((((((((((((..........))))))))))))..))).))))).......((((((((......)))))))).................. ( -34.10) >DroSim_CAF1 13751 120 - 1 CCGUCAAGCAAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGUCGAGGAGGUUUGAGAGCCAAUAAAGACGGGAACUAAUUAAUAAAUUUCCCUUUAUUAACUCAUACCUUUUAAGUUU (((((..((...((((....))))..))((((((((((((..........))))))))))))......))))).......((((((((......)))))))).................. ( -33.70) >consensus CCGUCAACCAAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGUCGAGGAGGUUUGAGAGCCUAUGAAGACGGGAACUAAUUAAUAAAUUUCCCUUUAUUAACUCAUACCUUUUAAGUUU (((((.............(((....)))((((((((((((...(....).))))))))))))......))))).......((((((((......)))))))).................. (-34.11 = -33.23 + -0.88)


| Location | 5,771,329 – 5,771,449 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -34.13 |
| Consensus MFE | -30.17 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5771329 120 + 23771897 UUAGUUACCGCCUUCAUAGGCUCUCAAACUUCCUCGGCACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUCGGUUGACGGCGCUUAAAAGCUCCCAAAGCUUCGCAUCGAGAAAACACCC ...(((...(((.(((..((((((((((((.(....)...))))))))))))..))).).)).....(((((((.(.(((.((((...........)))).))))))))))).))).... ( -35.60) >DroSec_CAF1 13928 112 + 1 UUAGUUCCCGUCUUCAUAGGCUCUCAAACCUCCUCGACACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUUGGUUGACGGCGCUUAAAAGCUCCCAAAGCUUCGCAUCGAGA-------- .......(((((.(((..(((((((((((............)))))))))))..)))..((((........))))))))).((....(((((.....))))).)).......-------- ( -33.40) >DroSim_CAF1 13791 112 + 1 UUAGUUCCCGUCUUUAUUGGCUCUCAAACCUCCUCGACACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUUGCUUGACGGCGCUUAAAAGCUCCCAAAGCUUCGCAUCGAGA-------- ..........((((....(((((((((((............)))))))))))((((.(((((.........))))).))))((....(((((.....))))).))...))))-------- ( -33.40) >consensus UUAGUUCCCGUCUUCAUAGGCUCUCAAACCUCCUCGACACAGUUUGAGAGCCGCUGACGAGCCCCCAUUUUGGUUGACGGCGCUUAAAAGCUCCCAAAGCUUCGCAUCGAGA________ .......(((((......(((((((((((............)))))))))))(((....))).............))))).((....(((((.....))))).))............... (-30.17 = -30.50 + 0.33)

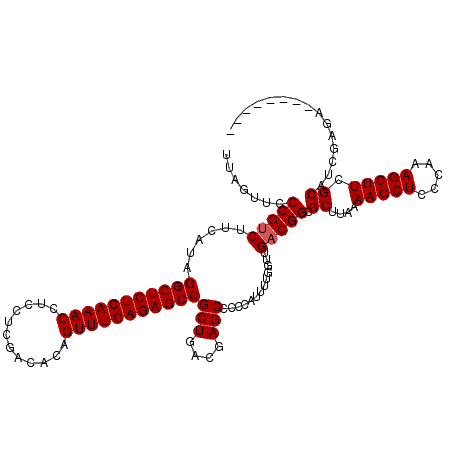
| Location | 5,771,329 – 5,771,449 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -39.47 |
| Energy contribution | -39.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5771329 120 - 23771897 GGGUGUUUUCUCGAUGCGAAGCUUUGGGAGCUUUUAAGCGCCGUCAACCGAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGCCGAGGAAGUUUGAGAGCCUAUGAAGGCGGUAACUAA .(((....((((((.((...)).))))))(((((((.(.(((.(((.......))).))).)......((((((((((((...(....).))))))))))))..)))))))....))).. ( -42.90) >DroSec_CAF1 13928 112 - 1 --------UCUCGAUGCGAAGCUUUGGGAGCUUUUAAGCGCCGUCAACCAAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGUCGAGGAGGUUUGAGAGCCUAUGAAGACGGGAACUAA --------(((((..(((((((((...)))))))...))(((.(((.......))).)))((.(((..((((((((((((..........))))))))))))..))).)))))))..... ( -41.10) >DroSim_CAF1 13791 112 - 1 --------UCUCGAUGCGAAGCUUUGGGAGCUUUUAAGCGCCGUCAAGCAAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGUCGAGGAGGUUUGAGAGCCAAUAAAGACGGGAACUAA --------(((((..(((((((((...)))))))...(.(((.(((.......))).))).)....))((((((((((((..........))))))))))))........)))))..... ( -40.70) >consensus ________UCUCGAUGCGAAGCUUUGGGAGCUUUUAAGCGCCGUCAACCAAAAUGGGGGCUCGUCAGCGGCUCUCAAACUGUGUCGAGGAGGUUUGAGAGCCUAUGAAGACGGGAACUAA ........(((((..(((((((((...)))))))...(.(((.(((.......))).))).)....))((((((((((((...(....).))))))))))))........)))))..... (-39.47 = -39.37 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:35 2006