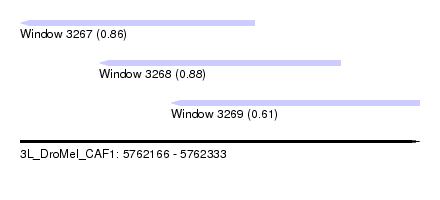
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,762,166 – 5,762,333 |
| Length | 167 |
| Max. P | 0.880091 |

| Location | 5,762,166 – 5,762,264 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
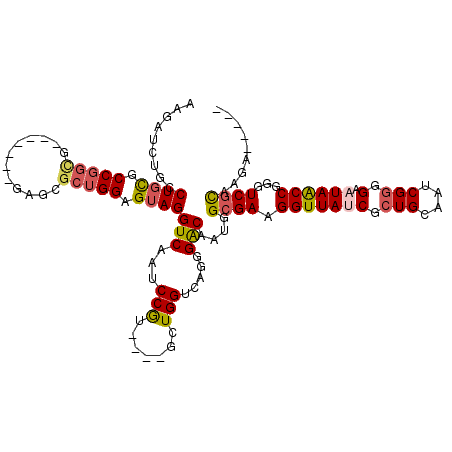
>3L_DroMel_CAF1 5762166 98 - 23771897 UAUCACUGGAAGCGGGGAAUAACCGAGUCGCAAGACUACAGGAGAUUAGCACACGGAAGUAUAC-CAAAGAAUGUAACCACUUUGGGUUACUACAAUUG------- ..((..(((..((....(((..((.((((....))))...))..))).))..((....))...)-))..))..((((((......))))))........------- ( -25.70) >DroSec_CAF1 4768 106 - 1 UAUCGCUGCAAUCGGGGAAUAACCGAGUCGCAAGACUACAGGAGAUACCAACACGGAAGUCUCCAUAAAGGAUGUAACCACUUUGGGUUAUACUGAUUGAUUUAAA ........(((((((.......((.((((....))))...((((((.((.....))..)))))).....)).(((((((......))))))))))))))....... ( -33.50) >DroSim_CAF1 4549 106 - 1 UAUCGCUGCAAUCGGGGAAUAACCGGGUCGUAAGACUACAGGAGAUACCAACACGGAAGUUUACAUAAAGGAUGUAACCACUUUGGGUUAAACUGAUUGAUUUAAG ........(((((((((.((..((.((((....))))...))..)).))(((.(.(((((((((((.....))))))..))))).))))...)))))))....... ( -29.20) >DroEre_CAF1 4718 105 - 1 UACCGCUGCAAGCGGGG-AUAGCCGGGUCGUAAGACUCCAGGAGACCCCAACACGGAAGUCUCCGUAAGGGAUGAAACCACUUUCAGUUUAAAUUAUGGAUUUAGG ..((((((.(((.((..-....(((((((....))).)).((((((.((.....))..)))))).....))......)).))).)))).((((((...)))))))) ( -32.20) >consensus UAUCGCUGCAAGCGGGGAAUAACCGAGUCGCAAGACUACAGGAGAUACCAACACGGAAGUCUACAUAAAGGAUGUAACCACUUUGGGUUAAAAUGAUUGAUUUAAG ..((.(((....(((.......)))((((....)))).)))...........((....)).........)).(((((((......))))))).............. (-17.74 = -17.68 + -0.06)


| Location | 5,762,199 – 5,762,300 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5762199 101 - 23771897 GGUCAAUCCGUGCUGGUCAGGGACAAUGGCGAAGGUUAUCACUGGAAGCGGGGAAUAACCGAGUCGCAAGACUACAGGAGAUUAGCACACGGAAGUAUAC-C ((.....))(((((((((........((((....))))((.(((....(((.......)))((((....)))).))))))))))))))((....))....-. ( -34.80) >DroSec_CAF1 4808 102 - 1 GGUCAAUCCGUGCUGGUCAGGGGCACCAGCGAAGGUUAUCGCUGCAAUCGGGGAAUAACCGAGUCGCAAGACUACAGGAGAUACCAACACGGAAGUCUCCAU ((....((((((((((((....).))))))...(((.(((.(((...((((.......))))(((....)))..)))..))))))...))))).....)).. ( -36.40) >DroSim_CAF1 4589 102 - 1 GGUCAAUCCGUGCUGGUCAGGGGCACCAGCGAAGGUUAUCGCUGCAAUCGGGGAAUAACCGGGUCGUAAGACUACAGGAGAUACCAACACGGAAGUUUACAU (((...(((.((((((((....).)))))))..(((((((.(((....))).).)))))).((((....))))...)))...)))...((....))...... ( -33.90) >DroEre_CAF1 4758 101 - 1 GGUCAAUCCGCGCUGGUCAGGGACAAUGGCGAAGGUUACCGCUGCAAGCGGGG-AUAGCCGGGUCGUAAGACUCCAGGAGACCCCAACACGGAAGUCUCCGU (((..((((.((((.(.(((((....((((....)))))).)))).)))).))-)).)))(((((....))).)).((((((.((.....))..)))))).. ( -43.40) >consensus GGUCAAUCCGUGCUGGUCAGGGACAACAGCGAAGGUUAUCGCUGCAAGCGGGGAAUAACCGAGUCGCAAGACUACAGGAGAUACCAACACGGAAGUCUACAU ((((..(((.(((((((.......)))))))..(((((((.(((....))).).)))))).((((....))))...))))))).....((....))...... (-27.25 = -27.12 + -0.12)

| Location | 5,762,229 – 5,762,333 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -23.96 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5762229 104 - 23771897 AAGAUCGGCCUGUGCCGGUG-------GAGCGCUGGAGUAGGUCAAUCCGU----GCUGGUCAGGGACAAUGGCGAAGGUUAUCACUGGAAGCGGGGAAUAACCGAGUCGCAAGA----- ......((((((..((((((-------...))))))..))))))..(((.(----(.....)).))).....((((.(((((((.(((....))).).))))))...))))....----- ( -37.80) >DroSec_CAF1 4839 104 - 1 AAGAUCUGCCUGCGCCGGCG-------GAGCGCUGGAGUAGGUCAAUCCGU----GCUGGUCAGGGGCACCAGCGAAGGUUAUCGCUGCAAUCGGGGAAUAACCGAGUCGCAAGA----- ..(((..((((((.((((((-------...)))))).))))))..))).((----(((.......)))))..((((.(((((((.(((....))).).))))))...))))....----- ( -42.60) >DroSim_CAF1 4620 104 - 1 AAGAUCUGCCUGCGCCGGCG-------GAGCGCUGGAGUAGGUCAAUCCGU----GCUGGUCAGGGGCACCAGCGAAGGUUAUCGCUGCAAUCGGGGAAUAACCGGGUCGUAAGA----- ..(((((((((((.((((((-------...)))))).)))))).......(----(((((((....).)))))))..(((((((.(((....))).).)))))))))))......----- ( -41.80) >DroEre_CAF1 4789 95 - 1 AAAAUGUGCCUGCUCCGGU---------------GGAGUAGGUCAAUCCGC----GCUGGUCAGGGACAAUGGCGAAGGUUACCGCUGCAAGCGGGG-AUAGCCGGGUCGUAAGA----- ....((.(((((((((...---------------)))))))))))(((((.----((((..(........((((....))))((((.....))))).-.))))))))).......----- ( -35.40) >DroAna_CAF1 5460 120 - 1 AAAAUGUGGCUCCACCGGCCCGGCCCGGCCAGCUGGAGUAGGUCAGUCCAUUGUCUCUGGUCAGGGACAAUGGCGAAGGUUAGCAGUGCAAUCGGGGAAUAACCGGGUCACCAAAAAUUA .....(((....))).((((((((((((((.((....)).)))).(.(((((((((((....))))))))))))...((((.((...)))))).))).....)))))))........... ( -45.40) >consensus AAGAUCUGCCUGCGCCGGCG_______GAGCGCUGGAGUAGGUCAAUCCGU____GCUGGUCAGGGACAAUGGCGAAGGUUAUCGCUGCAAUCGGGGAAUAACCGGGUCGCAAGA_____ .........((((.(((((............))))).))))(((...(((.......))).....)))....((((.(((((((.(((....))).).))))))...))))......... (-23.96 = -24.48 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:22 2006