| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 575,569 – 575,663 |
| Length | 94 |
| Max. P | 0.938890 |

| Location | 575,569 – 575,663 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
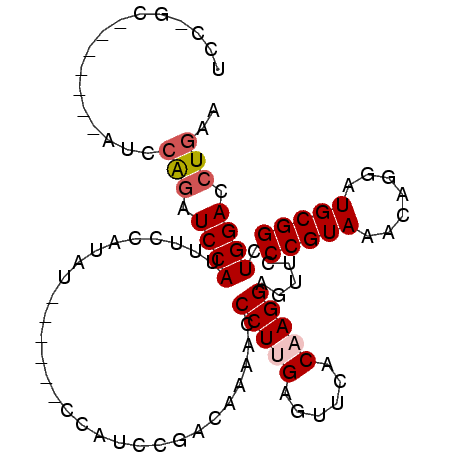
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -23.15 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 575569 94 + 23771897 UUCAGGUCCAGCCGCAUCCUGUUUACGGGAACUCCUUGUGAACUCAAGGGUUUUGUCGGAUGGAUGUGGAUAUGGAAAUGGAUCUGGAU-------GC-GGA ((((((((((.((((((((.((((((((.(((.(((((......)))))))))))).)))))))))))).........)))))))))).-------..-... ( -36.60) >DroSec_CAF1 6996 88 + 1 UUCAGGUCCAGCCGCAUCCUGUUUACGGGAACUCCUGGUAAACUCAAGGGUUUUGUCGGAUGG------AUAUGGAAAUGGAUCUUGAU-------GC-GGA .(((((((((.(((.((((.((((.((((....))))..(((((....)))))....))))))------)).)))...)))))).))).-------..-... ( -26.20) >DroSim_CAF1 7593 88 + 1 UUCAGGUCCAGCCGCAUCCUGUUUACGGUAACUCCUGGUGAACUCAAGGGUUUUGUCUGAUGG------AUAUGGAAAUGGAUCUGGAU-------GC-GGA ((((((((((.(((.((((.(((.((((.(((((.(((....).)).)))))))))..)))))------)).)))...)))))))))).-------..-... ( -30.50) >DroEre_CAF1 6877 96 + 1 UUCAGGUCCAGCCGCAUCCUGUUUACGGGAGUUCCUUGGGAACUCAAGGGUUUCCUCGGGUGG------AUGUGGAUAUGGAUGCGGAUGAAUGAUGCAGGG .(((..((...((((((((((((((((.....(((..(((((((....)))))))..)))...------.)))))))).))))))))..)).)))....... ( -38.40) >consensus UUCAGGUCCAGCCGCAUCCUGUUUACGGGAACUCCUGGUGAACUCAAGGGUUUUGUCGGAUGG______AUAUGGAAAUGGAUCUGGAU_______GC_GGA ((((((((((.((...(((((....)))))..(((....(((((....)))))....))).............))...)))))))))).............. (-23.15 = -24.02 + 0.88)

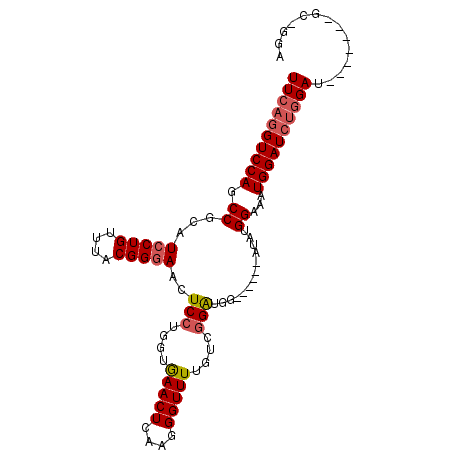
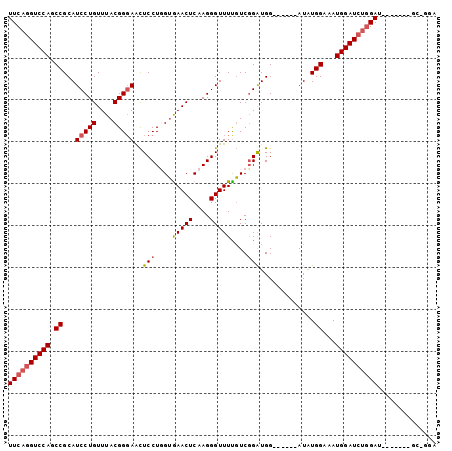
| Location | 575,569 – 575,663 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 575569 94 - 23771897 UCC-GC-------AUCCAGAUCCAUUUCCAUAUCCACAUCCAUCCGACAAAACCCUUGAGUUCACAAGGAGUUCCCGUAAACAGGAUGCGGCUGGACCUGAA ...-..-------...(((.((((.........((.(((((.....((..((((((((......))))).)))...)).....))))).)).)))).))).. ( -22.50) >DroSec_CAF1 6996 88 - 1 UCC-GC-------AUCAAGAUCCAUUUCCAUAU------CCAUCCGACAAAACCCUUGAGUUUACCAGGAGUUCCCGUAAACAGGAUGCGGCUGGACCUGAA ...-..-------.(((.(.((((...((.(((------((......(((.....))).((((((..((....)).)))))).))))).)).))))).))). ( -20.60) >DroSim_CAF1 7593 88 - 1 UCC-GC-------AUCCAGAUCCAUUUCCAUAU------CCAUCAGACAAAACCCUUGAGUUCACCAGGAGUUACCGUAAACAGGAUGCGGCUGGACCUGAA ...-..-------...(((.((((...((.(((------((.....((..(((.((((.(....))))).)))...)).....))))).)).)))).))).. ( -19.40) >DroEre_CAF1 6877 96 - 1 CCCUGCAUCAUUCAUCCGCAUCCAUAUCCACAU------CCACCCGAGGAAACCCUUGAGUUCCCAAGGAACUCCCGUAAACAGGAUGCGGCUGGACCUGAA .......(((((((.((((((((..........------......((((....))))(((((((...))))))).........)))))))).))))..))). ( -31.90) >consensus UCC_GC_______AUCCAGAUCCAUUUCCAUAU______CCAUCCGACAAAACCCUUGAGUUCACAAGGAGUUCCCGUAAACAGGAUGCGGCUGGACCUGAA ................(((.((((.............................(((((......))))).....(((((.......))))).)))).))).. (-13.69 = -14.50 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:13 2006