
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,740,489 – 5,740,609 |
| Length | 120 |
| Max. P | 0.705192 |

| Location | 5,740,489 – 5,740,609 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
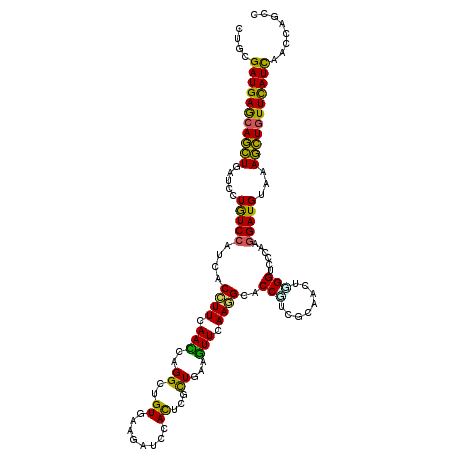
| Mean single sequence MFE | -34.81 |
| Consensus MFE | -21.79 |
| Energy contribution | -19.85 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
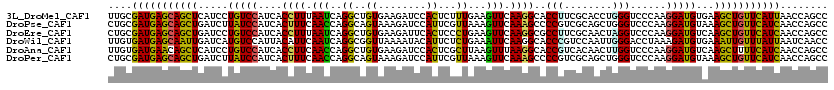
>3L_DroMel_CAF1 5740489 120 + 23771897 UUGCGAUGAGCAGCUCAUCCUGUCCAUCACCUUUAAUCAGGCUGUGAAGAUCCACUCUUUGAAGUUCAAGGCACCUUCGCACCUGGGUCCCAAGGAUGUGAAGCUGUUCAUUAACCAGCC ....(((((((((((((((((.......(((......((((.(((((((.......(((((.....)))))...))))))))))))))....))))))...)))))))))))........ ( -39.20) >DroPse_CAF1 21266 120 + 1 CUGCGAUGAGCAGCUGAUCUUAUCCAUCACUUUCAACCAGGCAGUAAAGAUCCAUUCGUUAAAGUUCAAAGCCCCGUCGCAGCUGGGUCCCAAGGAUGUAAAGCUGUUCAUCAACCAGCC ....(((((((((((.....(((((...(((.((.....)).)))...((((((((((.....((.....)).....)).)).))))))....)))))...)))))))))))........ ( -32.10) >DroEre_CAF1 10266 120 + 1 CUGCGAUGAGCAGCUGAUCCUGUCCAUCACCUUUAAUCAGGCUGUGAAGAUUCACUCCCUGAAGUUCAAGGCGCCUUCGCAACUAGGUCCCAAGGAUGUCAAGCUGUUCAUCAACCAGCC ....((((((((((((((...((((...........(((((..((((....))))..))))).......((.((((........)))).))..))))))).)))))))))))........ ( -43.90) >DroWil_CAF1 11900 120 + 1 UUGUGAUGAGCAAUUGAUCAUGUCCAUUACAUUCAAUCAGGCGGUUAAAAUACAUUCUCUGAAAUUCAAGGCACCCGUCCAAUUGGGACCUAAAGAUGUGAAAUUGUUUAUUAAUCAACC ...((((((((((((..(((((((............(((((..((........))..)))))......(((..((((......)))).)))...)))))))))))))))))))....... ( -27.70) >DroAna_CAF1 9871 120 + 1 UUGUGAUGAACAGCUCAUCCUGUCCAUCACCUUCAACCAGGCUGUGAAGAUCCACUCGCUUAAGUUUAAGGCACCGUCACAACUUGGUCCCAAGGAUGUCAAGCUUUUCAUCAACCAGCC ...(((((((.((((((((((..............((((((.(((((.((.....))((((.......))))....))))).))))))....))))))...)))).)))))))....... ( -33.87) >DroPer_CAF1 18416 120 + 1 CUGCGAUGAGCAGCUGAUCUUAUCCAUCACUUUCAACCAGGCAGUAAAGAUCCAUUCGUUAAAGUUCAAAGCCCCGUCGCAGCUGGGUCCCAAGGAUGUAAAGCUGUUCAUCAACCAGCC ....(((((((((((.....(((((...(((.((.....)).)))...((((((((((.....((.....)).....)).)).))))))....)))))...)))))))))))........ ( -32.10) >consensus CUGCGAUGAGCAGCUGAUCCUGUCCAUCACCUUCAACCAGGCUGUGAAGAUCCACUCGCUGAAGUUCAAGGCACCGUCGCAACUGGGUCCCAAGGAUGUAAAGCUGUUCAUCAACCAGCC ....(((((((((((.....(((((....((((.(((..((..((........))...))...))).))))..(((........)))......)))))...)))))))))))........ (-21.79 = -19.85 + -1.94)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:05 2006