
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,736,759 – 5,736,964 |
| Length | 205 |
| Max. P | 0.909586 |

| Location | 5,736,759 – 5,736,857 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.46 |
| Mean single sequence MFE | -16.48 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5736759 98 - 23771897 UCAGCAGUUGCCUCAGUGUGGCCUCUGUCUGAACAUGGCACUGUCACAAUCGUAUCCAAUCUAUUAACCUGUUUUCUUAUACUUAUUAAAGU--------UAAUUU- (((((((..(((.......)))..))).))))...((((...))))..............................................--------......- ( -14.00) >DroSec_CAF1 6591 106 - 1 UCAGCAGUUGCCUCAGUGUGGCCUCUGUCUGAACAUGGCACUGUCACAAUCGUGUCCAAUCUAUUAAUCUGUUUUCCUAUACUUAUUAAAGUAAAUGCAUUAAUUA- (((((((..(((.......)))..))).))))...((((((..........))).))).....................(((((....))))).............- ( -20.10) >DroSim_CAF1 6590 106 - 1 UCAGCAGUUGCCUCAGUGUGGCCUCUGUCUGAACAUGGCACUGUCACAAUCGUGUCCAAUCUAUUAACCUGUUUUCCUAUACUUAUUAAAGUAAAUGCAUUAAUUA- (((((((..(((.......)))..))).))))...((((((..........))).))).....................(((((....))))).............- ( -20.10) >DroEre_CAF1 6546 86 - 1 UCAGCAGUUGCCUCUGUGUGGCCUCUGUCUGAACAUGGCACUGUCACAAUUCAAUCCA-CCUAUCAACCAAAUUUCACA--------------------UAAAGUAU (((((((..(((.......)))..))).))))...((((...))))............-....................--------------------........ ( -14.00) >DroYak_CAF1 6595 81 - 1 UCAGCAGUUGCCUUAGUGUGGCCUCUGUCUGAACAUGGCACUGUCACAAUUGAAACCA-CCUAUCUA----UUUAUGCA--------------------UUAAUUA- (((((((..(((.......)))..))).))))...(((.....(((....)))..)))-........----........--------------------.......- ( -14.80) >DroAna_CAF1 6737 78 - 1 UCAGCAGUUGCCGUUCUGUGGCCUCUGUCUGAACAUGCCACUGCUACAGU---AUACUUUAAAUGAAUCUUCCUUCCGCCA-------------------------- ..((((((.((.((((...(((....))).))))..)).)))))).....---............................-------------------------- ( -15.90) >consensus UCAGCAGUUGCCUCAGUGUGGCCUCUGUCUGAACAUGGCACUGUCACAAUCGUAUCCAAUCUAUUAACCUGUUUUCCUAUA__________________UUAAUUA_ (((((((..(((.......)))..))).))))........................................................................... (-13.15 = -13.15 + 0.00)



| Location | 5,736,789 – 5,736,893 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.43 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -24.10 |
| Energy contribution | -23.93 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5736789 104 - 23771897 ACAUUUUGCCAGAACAAAUUCC---AGGCAUUGUCGUCUUCAGCAGUUGCCUCAGUG--UGGCCUCUGUCUGAACAUGGCACUGUCACAAUCGUAUCCAAUCUAUUA--AC ......................---.((((.((((((.((((((((..(((......--.)))..))).))))).)))))).)))).....................--.. ( -27.40) >DroSec_CAF1 6629 104 - 1 ACAUUUUGCCAGAACAAAUCCC---AGGCAUUGUCGUCUUCAGCAGUUGCCUCAGUG--UGGCCUCUGUCUGAACAUGGCACUGUCACAAUCGUGUCCAAUCUAUUA--AU ...........(.(((......---.((((.((((((.((((((((..(((......--.)))..))).))))).)))))).)))).......))).).........--.. ( -28.14) >DroEre_CAF1 6565 103 - 1 ACAUUUUGCCAGAACAAAUCUC---AGGCUUUGACGUCUUCAGCAGUUGCCUCUGUG--UGGCCUCUGUCUGAACAUGGCACUGUCACAAUUCAAUCCA-CCUAUCA--AC ......(((((...........---((((......))))(((((((..(((......--.)))..))).))))...)))))..................-.......--.. ( -22.60) >DroYak_CAF1 6610 102 - 1 ACAUUUUGCCAGAACAAAUCCC---AGGCUUUGACGUCUUCAGCAGUUGCCUUAGUG--UGGCCUCUGUCUGAACAUGGCACUGUCACAAUUGAAACCA-CCUAUCU--A- ......(((((...((((.(..---..).)))).....((((((((..(((......--.)))..))).)))))..)))))..................-.......--.- ( -22.80) >DroMoj_CAF1 6854 110 - 1 ACAUUUUGCUAGAAUAAAGCUCCAAAGGCUUUGGCGUCUUCAGCAGUUGCCUCAUUUCAUGGCCUCGGUCUGAACAUGCCAUUGCUACAAAAGUUCCUAUA-UAUUCCCAG ...(((((.(((...((((((.....))))))(((((.((((((.(..(((.........)))..).).))))).)))))....)))))))).........-......... ( -26.60) >DroAna_CAF1 6750 101 - 1 ACAUUCUGCCAGACCAAAGCCC---AGGCAUUGGCGUCUUCAGCAGUUGCCGUUCUG--UGGCCUCUGUCUGAACAUGCCACUGCUACAGU---AUACUUUAAAUGA--AU ......................---.((((.((((((.((((((((..((((.....--))))..))).))))).)))))).)))).....---.............--.. ( -30.30) >consensus ACAUUUUGCCAGAACAAAUCCC___AGGCAUUGACGUCUUCAGCAGUUGCCUCAGUG__UGGCCUCUGUCUGAACAUGGCACUGUCACAAUCGUAUCCA_UCUAUUA__AC ..........................(((..((((((.((((((((..(((.........)))..))).))))).))))))..)))......................... (-24.10 = -23.93 + -0.17)



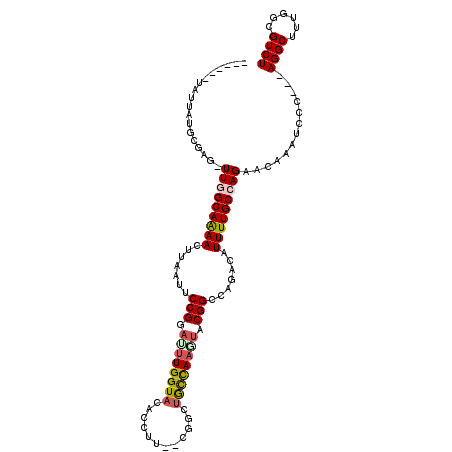
| Location | 5,736,857 – 5,736,964 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.90 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -18.80 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5736857 107 - 23771897 AUUCCCUAUUAUGCGAG-UUGGCAGAACUUAAUUCCGGACCUGGUACACCUU--CGGGUGCUAAGUGCGGCCAGACAUUUUGCCAGAACAAAUUCC---AGGCAUUGUCGUCU ((((.(......).)))-)((((...(((((...(((....)))..((((..--..)))).)))))...))))((((...((((.(((....))).---.)))).)))).... ( -30.20) >DroVir_CAF1 6820 107 - 1 ------UAUGAUACUAGUUUUGCAAAACUUAAUUCCGGAUUUGGUAUAACUCUUGAGCUGCCAACUACGGCCAGACAUUUUGCUAGAACAAAGCUCCAAAGGCUUUGGCGUCU ------...((..((.(((((((((((.......(((...((((((...((....)).))))))...))).......)))))).)))))((((((.....))))))))..)). ( -24.54) >DroGri_CAF1 6998 107 - 1 ------UAUGAUUUGAUUUUUGCAAAACUUAAUUCCGGAUUUGGUAUAACUCUUGAGCUGCCAACUACGGCCAGACAUUUUGCUAGAGCAAAGCUCCAAAGGCUUUGGCGUCU ------...(((....((((.((((((.......(((...((((((...((....)).))))))...))).......)))))).))))(((((((.....)))))))..))). ( -22.44) >DroEre_CAF1 6632 101 - 1 ------UAAUACGCUAG-UUGGCAGAACUUAAUUCCGGAUUUGGUACACCUU--CGGGUGCUAAGUACGGCCAGACAUUUUGCCAGAACAAAUCUC---AGGCUUUGACGUCU ------...........-(((((((((.......(((.((((((..((((..--..)))))))))).))).......)))))))))..........---((((......)))) ( -27.74) >DroYak_CAF1 6676 101 - 1 ------UAUUAUGCGAG-UUGGCAGAACUUAAUUCCGGAUUUGGUACACCUU--CGGGUGCUAAGUACGGCCAGACAUUUUGCCAGAACAAAUCCC---AGGCUUUGACGUCU ------......(((..-(((((((((.......(((.((((((..((((..--..)))))))))).))).......)))))))))..((((.(..---..).)))).))).. ( -29.24) >DroPer_CAF1 15123 92 - 1 ----------------A-UUGGCAAAACUUAAUUCCGGAUUUGGUUCACCUUU-UGGCUAUCAAACACGGCCAGACAUUUUGCCAGAGCAAACCCC---AGGCCUUGGCGUCU ----------------.-(((((((((.......(((..((((((...((...-.))..))))))..))).......)))))))))..........---..((....)).... ( -23.64) >consensus ______UAUUAUGCGAG_UUGGCAAAACUUAAUUCCGGAUUUGGUACACCUU__CGGCUGCCAAGUACGGCCAGACAUUUUGCCAGAACAAAUCCC___AGGCUUUGGCGUCU ..................(((((((((.......(((.((((((((............)))))))).))).......))))))))).............((((......)))) (-18.80 = -19.02 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:03 2006