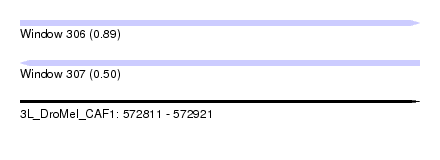
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 572,811 – 572,921 |
| Length | 110 |
| Max. P | 0.885906 |

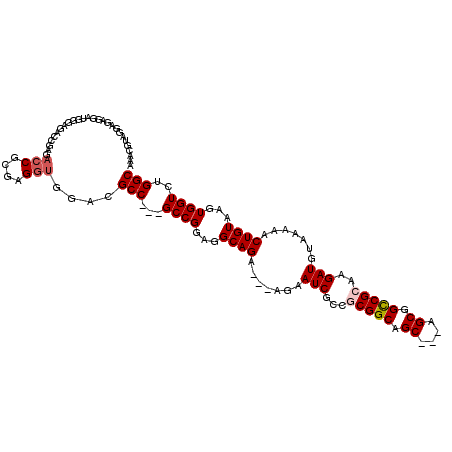
| Location | 572,811 – 572,921 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -24.69 |
| Energy contribution | -27.00 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 572811 110 + 23771897 GCCAGACCACUUACAGUUUUUACAUCUUGCGACCGCU---GCCGCCGCGGCGAUUCU---UCUGCCUCCGGC---GGCGUCCACCUCGCGGUCUCGGUCUCCCAUCCUCUCCUACAUUU ...(((((.......((....)).....(.(((((((---(((((((.(((((....---)).)))..))))---))))........)))))).))))))................... ( -37.70) >DroSec_CAF1 4232 110 + 1 GCCAGACCACUUACAGUUUUUACAUCUUGCGGCCGCU---GCUGCCGAGGCGAUUCU---UCUGCCUCCGGC---GGCGUCCACCUCGCGGUCUCGGUCUCCCAUCCCCUCGUACAUUU ...(((((.......((....)).....(.(((((((---(((((((((((((....---)).)))).))))---))))........)))))).))))))................... ( -37.50) >DroSim_CAF1 4657 110 + 1 GCCAGACCACUUACAGUUUUUACAUCUUGCGGCCGCU---GCUGCCGAGGCGAUUCU---UCUGCCUCCGGC---GGCGUUCACCUCGCGGUCUCGUUCUCCCAUCCUCUCCUACGUUU ((.(((((.......((....)).....((((....(---(((((((((((((....---)).)))).))))---))))......))))))))).))...................... ( -33.10) >DroEre_CAF1 4176 112 + 1 GCCACACCACUUACAGUUUUUACAUCUUGCGGCCGCCGACGCUGCCGCGGCGAUUCUUCUUCUGCCUCCGGCGGCGGCGUCCACCUCGCG-------CCUCCCACCCUCUCCUACGUGC ..(((..........((....)).......(((((..((((((((((((((((.......)).)))....))))))))))).....)).)-------))................))). ( -33.80) >consensus GCCAGACCACUUACAGUUUUUACAUCUUGCGGCCGCU___GCUGCCGAGGCGAUUCU___UCUGCCUCCGGC___GGCGUCCACCUCGCGGUCUCGGUCUCCCAUCCUCUCCUACAUUU ((((((((.......((....)).....((((.(((((((((((..((((((..........)))))))))))))))))......))))))))).)))..................... (-24.69 = -27.00 + 2.31)

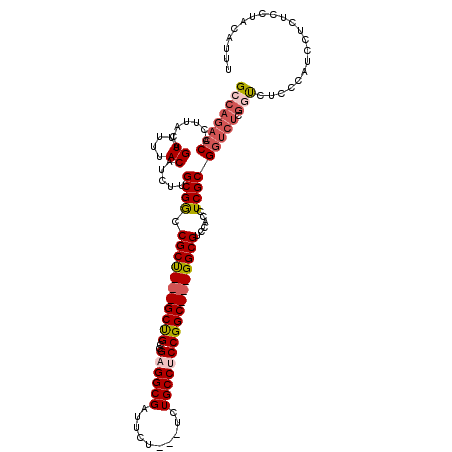
| Location | 572,811 – 572,921 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -39.53 |
| Consensus MFE | -26.63 |
| Energy contribution | -27.44 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 572811 110 - 23771897 AAAUGUAGGAGAGGAUGGGAGACCGAGACCGCGAGGUGGACGCC---GCCGGAGGCAGA---AGAAUCGCCGCGGCGGC---AGCGGUCGCAAGAUGUAAAAACUGUAAGUGGUCUGGC ...............(((....)))(((((((..(((..(((((---((((..(((.((---....))))).)))))))---....(((....)))))....)))....)))))))... ( -45.30) >DroSec_CAF1 4232 110 - 1 AAAUGUACGAGGGGAUGGGAGACCGAGACCGCGAGGUGGACGCC---GCCGGAGGCAGA---AGAAUCGCCUCGGCAGC---AGCGGCCGCAAGAUGUAAAAACUGUAAGUGGUCUGGC ...............(((....)))(((((((..(((.(..((.---((((.((((.((---....)))))))))).))---..).)))((.....))...........)))))))... ( -39.50) >DroSim_CAF1 4657 110 - 1 AAACGUAGGAGAGGAUGGGAGAACGAGACCGCGAGGUGAACGCC---GCCGGAGGCAGA---AGAAUCGCCUCGGCAGC---AGCGGCCGCAAGAUGUAAAAACUGUAAGUGGUCUGGC .......................(.(((((((..(((....((.---((((.((((.((---....)))))))))).))---.(((..(....).)))....)))....))))))).). ( -34.30) >DroEre_CAF1 4176 112 - 1 GCACGUAGGAGAGGGUGGGAGG-------CGCGAGGUGGACGCCGCCGCCGGAGGCAGAAGAAGAAUCGCCGCGGCAGCGUCGGCGGCCGCAAGAUGUAAAAACUGUAAGUGGUGUGGC ((((.........(((((..((-------(((......).)))).)))))....((((.......(((...(((((.((....)).)))))..))).......)))).....))))... ( -39.04) >consensus AAACGUAGGAGAGGAUGGGAGACCGAGACCGCGAGGUGGACGCC___GCCGGAGGCAGA___AGAAUCGCCGCGGCAGC___AGCGGCCGCAAGAUGUAAAAACUGUAAGUGGUCUGGC ...........................(((....)))....(((...((((...((((.......(((...(((((.((....)).)))))..))).......))))...))))..))) (-26.63 = -27.44 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:09 2006