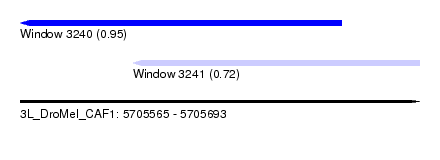
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,705,565 – 5,705,693 |
| Length | 128 |
| Max. P | 0.951524 |

| Location | 5,705,565 – 5,705,668 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5705565 103 - 23771897 CUUCAGGAGGUCAUCCUCCUGA-------CACCAACCUCCCUCCACCUUCCUCCUCGUGAUCAUACCAACGAUUUUCGGGCACUGCACUUACUUGCUCCGCGGUCUUUAG ..((((((((....))))))))-------.(((.....................(((.((((........))))..)))((...(((......)))...)))))...... ( -23.70) >DroSec_CAF1 79336 100 - 1 CUUCAGGAGGUCAUCCUCCUGC-------CACCAACCUCCCUCCACCUUCCUCC---UGAUCAUACCAACGAUUUUCGGGCACUGCACUUACUUGCUCCGCGGUCUUUAG ...(((((((....))))))).-------.(((...................((---.((((........))))...))((...(((......)))...)))))...... ( -21.30) >DroSim_CAF1 78708 100 - 1 CUUCAGGAGGUCAUCCUCCUGC-------CACCAACCUCCCUCCACCUUCCUCC---UGAUCAUACCAACGAUUUUCGGGCACUGCACUUACUUGCUCCGCGGUCUUUAG ...(((((((....))))))).-------.(((...................((---.((((........))))...))((...(((......)))...)))))...... ( -21.30) >DroEre_CAF1 81580 100 - 1 CUUCAGGAGGUCAUCCUCCAGC-------CGCCAACCUCCCUCCACCUUCCUCC---UGAUCAUAACAACGAUUUUCGGGCACUGCACUUACUUGCUCCGCGGUCUUUAG .....(((((....))))).((-------(((.................(((..---.((((........))))...)))....(((......)))...)))))...... ( -22.30) >DroYak_CAF1 82328 107 - 1 CUUCAGGAGGUCAUCCUCCUGCCUUGUGCCUCCAACCUCCCUCCACCUUCCUCC---AGAUCAUACAAACGUGUUUCGGACGAUGCACUUACUUGCUCCGCGGUCUUUAG ...(((((((....)))))))......(((.....................(((---.((..((((....)))).)))))((..(((......)))..)).)))...... ( -21.60) >consensus CUUCAGGAGGUCAUCCUCCUGC_______CACCAACCUCCCUCCACCUUCCUCC___UGAUCAUACCAACGAUUUUCGGGCACUGCACUUACUUGCUCCGCGGUCUUUAG ...(((((((....))))))).....................................((((.......((.....)).((...(((......)))...))))))..... (-18.10 = -18.50 + 0.40)


| Location | 5,705,601 – 5,705,693 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5705601 92 - 23771897 GUUGUCCCCAUUCCAAGUUUUCCAGCUUCAGGAGGUCAUCCUCCUGA-------CACCAACCUCCCUCCACCUUCCUCCUCGUGAUCAUACCAACGAUU ((((...........(((......)))((((((((....))))))))-------...))))..................((((..........)))).. ( -17.90) >DroEre_CAF1 81616 89 - 1 GUUGUCCCCAUUCCCAGUUGUCCAGCUUCAGGAGGUCAUCCUCCAGC-------CGCCAACCUCCCUCCACCUUCCUCC---UGAUCAUAACAACGAUU ................(((((......((((((((............-------....................)))))---))).....))))).... ( -16.95) >DroYak_CAF1 82364 96 - 1 GUUGUCCCCAUUCCCAGUUUUCCAGCUUCAGGAGGUCAUCCUCCUGCCUUGUGCCUCCAACCUCCCUCCACCUUCCUCC---AGAUCAUACAAACGUGU ((((....(((....(((......))).(((((((....)))))))....)))....))))..................---.....((((....)))) ( -14.10) >consensus GUUGUCCCCAUUCCCAGUUUUCCAGCUUCAGGAGGUCAUCCUCCUGC_______CACCAACCUCCCUCCACCUUCCUCC___UGAUCAUACCAACGAUU ((((...........((((....)))).(((((((....)))))))...........))))...................................... (-12.47 = -12.80 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:55 2006