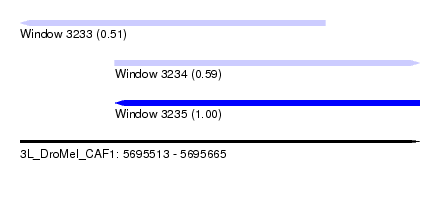
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,695,513 – 5,695,665 |
| Length | 152 |
| Max. P | 0.999045 |

| Location | 5,695,513 – 5,695,629 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -19.29 |
| Consensus MFE | -12.01 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5695513 116 - 23771897 AGAGUCUUUCUCUUUUAAGUAAAAACUAGACAGGGCGCACAGAAAAUGCACACAGCUCCUGUCUACUUUCAAUUUAAACUUAGUCUUUUUUAAUCCUAUUUGCAAAGCCAGCUUAA ((((.....)))).((((((......(((((((((((((.......))).....)).))))))))(((((((.......((((......))))......))).))))...)))))) ( -20.52) >DroSec_CAF1 68098 103 - 1 AGAGUUUUUCUCUUUUAAGUAAAAACUAGACAGGG-------------CACACAGCUCCUGUCUACUUUCAAUUUAAACUUAAUCUUUUUUAAUCCUAUUUGCAAAGCCAGCUUAA ((((.....)))).((((((......(((((((((-------------(.....)).))))))))(((((((.......((((......))))......))).))))...)))))) ( -21.02) >DroSim_CAF1 68419 103 - 1 AGAGUUUUUCUCUUUUAAGUAAAAACUAGACAGGG-------------CACACAGCUUCUGUCUACUUUCAAUUUAAACUUAAUCUUUUUUAAUCCUAUUUGCAAAGCCAGCUUAA ((((.....)))).((((((......(((((((((-------------(.....)).))))))))(((((((.......((((......))))......))).))))...)))))) ( -18.02) >DroEre_CAF1 71434 116 - 1 AGGGCUUUUCUUUUUUAAGUGAACUCUAAGCAGGGCAUACAUAAAAUGCACACCACUCUGGUCCACUUUCAAUUUAAAGUUAAUCUUUUUUAAUCCUAUUUGCACAGCCAGCCUUG ((((((......(((((.(((..((((....))))..))).)))))((((.(((.....)))..(((((......)))))....................)))).....)))))). ( -17.60) >consensus AGAGUUUUUCUCUUUUAAGUAAAAACUAGACAGGG_____________CACACAGCUCCUGUCUACUUUCAAUUUAAACUUAAUCUUUUUUAAUCCUAUUUGCAAAGCCAGCUUAA .(((((.......(((((((..(((.(((((((((.....................))))))))).)))..))))))).((((......))))................))))).. (-12.01 = -12.20 + 0.19)



| Location | 5,695,549 – 5,695,665 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -12.49 |
| Energy contribution | -13.55 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5695549 116 + 23771897 AGUUUAAAUUGAAAGUAGACAGGAGCUGUGUGCAUUUUCUGUGCGCCCUGUCUAGUUUUUACUUAAAAGAGAAAGACUCUUUCAAAGUACUCUCAACAAAUUUUCACACCGAA---UGU .((..(((((((((.((((((((....(((..((.....))..))))))))))).))))(((((.((((((.....))))))..))))).........)))))..))......---... ( -29.70) >DroSec_CAF1 68134 105 + 1 AGUUUAAAUUGAAAGUAGACAGGAGCUGUGUG-------------CCCUGUCUAGUUUUUACUUAAAAGAGAAAAACUCUUUCAAAGUACUCUCAACA-AUUUUCACACCGAAUAGUGU .((..(((((((((.((((((((.((.....)-------------))))))))).))).(((((.((((((.....))))))..))))).......))-))))..))............ ( -29.10) >DroSim_CAF1 68455 105 + 1 AGUUUAAAUUGAAAGUAGACAGAAGCUGUGUG-------------CCCUGUCUAGUUUUUACUUAAAAGAGAAAAACUCUUUCAAAGUACUCUCAACA-AUUUUCACACCGAAUAGUGU .((..(((((((((.(((((((..((.....)-------------).))))))).))).(((((.((((((.....))))))..))))).......))-))))..))............ ( -24.00) >DroEre_CAF1 71470 110 + 1 ACUUUAAAUUGAAAGUGGACCAGAGUGGUGUGCAUUUUAUGUAUGCCCUGCUUAGAGUUCACUUAAAAAAGAAAAGCCCUUUCAAAGUAC------CU-UUUUUCACACCGCAUAG--U (((((.......(((((((((.(((((((((((((...)))))))))..)))).).))))))))......((((.....)))))))))..------..-.................--. ( -24.40) >consensus AGUUUAAAUUGAAAGUAGACAGGAGCUGUGUG_____________CCCUGUCUAGUUUUUACUUAAAAGAGAAAAACUCUUUCAAAGUACUCUCAACA_AUUUUCACACCGAAUAGUGU ........(((((((((((((((.......................))))))))((((((.(((....))))))))).))))))).................................. (-12.49 = -13.55 + 1.06)

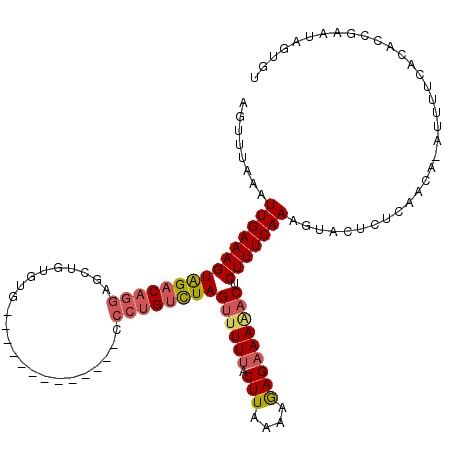
| Location | 5,695,549 – 5,695,665 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -18.40 |
| Energy contribution | -19.52 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5695549 116 - 23771897 ACA---UUCGGUGUGAAAAUUUGUUGAGAGUACUUUGAAAGAGUCUUUCUCUUUUAAGUAAAAACUAGACAGGGCGCACAGAAAAUGCACACAGCUCCUGUCUACUUUCAAUUUAAACU .((---(.....))).......(((((((((((((.(((((((.....)))))))))))).....(((((((((((((.......))).....)).))))))))))))))))....... ( -31.40) >DroSec_CAF1 68134 105 - 1 ACACUAUUCGGUGUGAAAAU-UGUUGAGAGUACUUUGAAAGAGUUUUUCUCUUUUAAGUAAAAACUAGACAGGG-------------CACACAGCUCCUGUCUACUUUCAAUUUAAACU (((((....)))))......-.(((((((((((((.(((((((.....)))))))))))).....(((((((((-------------(.....)).))))))))))))))))....... ( -34.80) >DroSim_CAF1 68455 105 - 1 ACACUAUUCGGUGUGAAAAU-UGUUGAGAGUACUUUGAAAGAGUUUUUCUCUUUUAAGUAAAAACUAGACAGGG-------------CACACAGCUUCUGUCUACUUUCAAUUUAAACU (((((....)))))......-.(((((((((((((.(((((((.....)))))))))))).....(((((((((-------------(.....)).))))))))))))))))....... ( -31.80) >DroEre_CAF1 71470 110 - 1 A--CUAUGCGGUGUGAAAAA-AG------GUACUUUGAAAGGGCUUUUCUUUUUUAAGUGAACUCUAAGCAGGGCAUACAUAAAAUGCACACCACUCUGGUCCACUUUCAAUUUAAAGU (--(((.(.((((((.....-..------((((((((..((((.(((.(((....))).)))))))...)))))..)))........)))))).)..))))..(((((......))))) ( -20.96) >consensus ACACUAUUCGGUGUGAAAAU_UGUUGAGAGUACUUUGAAAGAGUUUUUCUCUUUUAAGUAAAAACUAGACAGGG_____________CACACAGCUCCUGUCUACUUUCAAUUUAAACU ......................(((((((((((((.(((((((.....)))))))))))).....(((((((((.....................)))))))))))))))))....... (-18.40 = -19.52 + 1.13)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:49 2006