
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,689,764 – 5,689,854 |
| Length | 90 |
| Max. P | 0.638247 |

| Location | 5,689,764 – 5,689,854 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5689764 90 - 23771897 ----G---CUGCUGCCGCUGCGGU----GGCUGCCAUUUGACGAAAAUACAUAAAUCACGCGUCAUGUGCAGCUAAAAUUUUCCAGCUUU-CCCCAUCGCU----A--- ----(---(((((((....))))(----((((((.((.(((((.................))))).)))))))))........))))...-..........----.--- ( -23.73) >DroPse_CAF1 66764 97 - 1 GCUG----CUGCCUCUGAUGCGGU----GGCUGCCAUUUGACGAAAAUACAUAAAUCACGCGUCAUGUGCAGCUAAAAUUUUCCCCAUUUUCCCUUUUGCU----GGUG ((.(----(..((.(....).)).----.)).))(((.(((((.................))))).)))((((.........................)))----)... ( -20.74) >DroSec_CAF1 62395 90 - 1 ----G---CUGCUGCCGCUGCGGU----GGCUGCCAUUUGACGAAAAUACAUAAAUCACGCGUCAUGUGCAGCUAAAAUUUUGCAGCUUU-CCCCAUCGCU----C--- ----(---((((.((((...))))----((((((.((.(((((.................))))).))))))))........)))))...-..........----.--- ( -26.53) >DroSim_CAF1 62649 90 - 1 ----G---CUGCUGCCGCUGCGGU----GGCUGCCAUUUGACGAAAAUACAUAAAUCACGCGUCAUGUGCAGCUAAAAUUUUGCAGCUUU-CCCCCUCGCU----C--- ----(---((((.((((...))))----((((((.((.(((((.................))))).))))))))........)))))...-..........----.--- ( -26.53) >DroYak_CAF1 64523 78 - 1 ----GCUGCUGCUGCUGCUGCGGU----GGCUGCCAUUUGACGAAAAUACAUAAAUCACGCGUCAUGUGCAGCU---------------U-UCCCAAUGCA----C--- ----(((((.((....)).)))))----((((((.((.(((((.................))))).))))))))---------------.-..........----.--- ( -23.23) >DroAna_CAF1 63499 96 - 1 ----G------CUGCCACUGCGGUGGCUGGCUGCCAUUUGACGAAAAUACAUAAAUCACGCGUCAUGUGCAGCUAAAAUUGUUCCCCUCUACCCCGUUGCACAGAC--- ----(------(.(((((....)))))(((((((.((.(((((.................))))).))))))))).......................))......--- ( -25.93) >consensus ____G___CUGCUGCCGCUGCGGU____GGCUGCCAUUUGACGAAAAUACAUAAAUCACGCGUCAUGUGCAGCUAAAAUUUUCCAGCUUU_CCCCAUCGCU____C___ ..........(((((....)))))....((((((.((.(((((.................))))).))))))))................................... (-18.00 = -18.20 + 0.19)

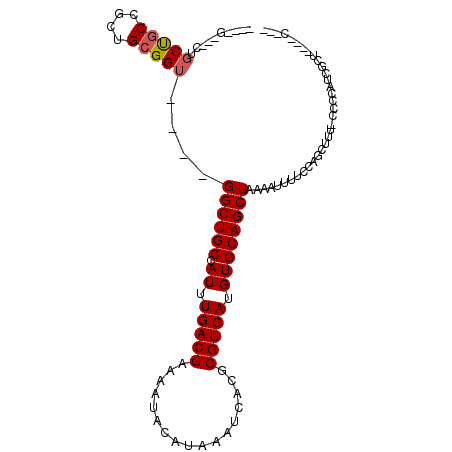

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:44 2006