
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,689,302 – 5,689,435 |
| Length | 133 |
| Max. P | 0.923959 |

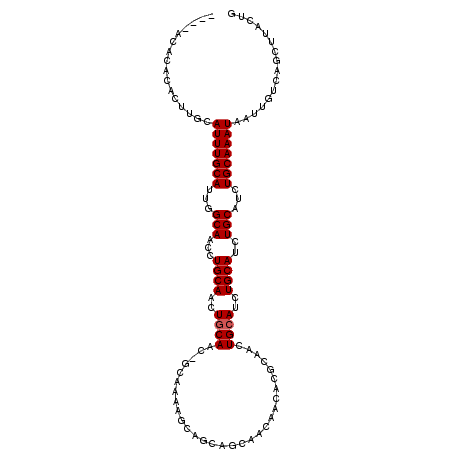
| Location | 5,689,302 – 5,689,415 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.23 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -14.71 |
| Energy contribution | -15.04 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5689302 113 + 23771897 ACACACACACACUUGUAUUUGCAUUGGCAACCUGCAACUGCAACAGCAAAAGCAGCAGCAACAACACGCAACUGCAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUG ..........((...((((((((...(((...((((..(((....)))...((((..((........))..))))...))))..)))...))))))))...)).......... ( -26.10) >DroEre_CAF1 65469 108 + 1 ----ACACAUACUUGCAUUUGCAUUGGCAACCUGCAACUGCAAC-GCAAAAGCAGCAACAACAACACACAACUGAAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUG ----........(((((.(((((..(....).))))).))))).-(((...((((...((............))...))))...)))..((((((....))).)))....... ( -20.00) >DroYak_CAF1 64055 97 + 1 ----ACACGCACUUACAUUUGCAUUGGCAACCUGCAACUGCAAC------------AGCAACAACACGCAACUGCAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUG ----....((((....(((((((...(((...((((..((((..------------.((........))...))))..))))..)))...)))))))....))..))...... ( -21.70) >consensus ____ACACACACUUGCAUUUGCAUUGGCAACCUGCAACUGCAAC_GCAAAAGCAGCAGCAACAACACGCAACUGCAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUG ................(((((((...(((...((((..((((..............................))))..))))..)))...)))))))................ (-14.71 = -15.04 + 0.33)

| Location | 5,689,302 – 5,689,415 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.23 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -27.49 |
| Energy contribution | -27.49 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5689302 113 - 23771897 CAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUGCAGUUGCGUGUUGUUGCUGCUGCUUUUGCUGUUGCAGUUGCAGGUUGCCAAUGCAAAUACAAGUGUGUGUGUGU ..((((.(((.(((((((((((....))))))(((((.(((((.(((......))).))))).)))))......)))))))).))))..(((((.((((....)))).))))) ( -37.80) >DroEre_CAF1 65469 108 - 1 CAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUUCAGUUGUGUGUUGUUGUUGCUGCUUUUGC-GUUGCAGUUGCAGGUUGCCAAUGCAAAUGCAAGUAUGUGU---- (((....)))(((..((((((((((((((((.((((...(((..........)))...)))).)))))-)))(((...((.....))...)))...))))))))..)))---- ( -31.80) >DroYak_CAF1 64055 97 - 1 CAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUGCAGUUGCGUGUUGUUGCU------------GUUGCAGUUGCAGGUUGCCAAUGCAAAUGUAAGUGCGUGU---- (((....)))(((..((((((((..((((...((((.((((..((((.((....)).------------..))))..))))..))))...))))..))))))))..)))---- ( -32.40) >consensus CAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUGCAGUUGCGUGUUGUUGCUGCUGCUUUUGC_GUUGCAGUUGCAGGUUGCCAAUGCAAAUGCAAGUGUGUGU____ (((....)))(((..((((((((..((((...((((.((((..((((........................))))..))))..))))...))))..))))))))..))).... (-27.49 = -27.49 + 0.01)



| Location | 5,689,335 – 5,689,435 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -16.53 |
| Energy contribution | -16.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5689335 100 + 23771897 GCAACUGCAACAGCAAAAGCAGCAGCAACAACACGCAACUGCAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUGACUCGCAUUCAAUGCAGCAA (((..((((...(((...((((..((........))..))))...)))...))))..)))........(((((....)))))..((((...))))..... ( -26.30) >DroEre_CAF1 65498 99 + 1 GCAACUGCAAC-GCAAAAGCAGCAACAACAACACACAACUGAAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUGACUCGCAUUCAACGCAGCAA ....((((...-((....))...................((((..(((...(((....))).......(((((....)))))..)))))))..))))... ( -19.70) >DroYak_CAF1 64084 88 + 1 GCAACUGCAAC------------AGCAACAACACGCAACUGCAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUGACUCGCAUUCAAUGCAGCAA (((..(((...------------.))).......((...((((..(((....)))..)))).......(((((....)))))..))......)))..... ( -20.80) >consensus GCAACUGCAAC_GCAAAAGCAGCAGCAACAACACGCAACUGCAUCUGCAUCUGCAUCUGCAAAUAAUUGUCAGCUUACUGACUCGCAUUCAAUGCAGCAA ....((((...............................((((..(((....)))..)))).......(((((....)))))...........))))... (-16.53 = -16.87 + 0.33)


| Location | 5,689,335 – 5,689,435 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.77 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5689335 100 - 23771897 UUGCUGCAUUGAAUGCGAGUCAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUGCAGUUGCGUGUUGUUGCUGCUGCUUUUGCUGUUGCAGUUGC ...(((((...(((....(((((....))))).)))...))))).(((((..(((((.(((((.(((......))).))))).)))))..)))))..... ( -36.70) >DroEre_CAF1 65498 99 - 1 UUGCUGCGUUGAAUGCGAGUCAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUUCAGUUGUGUGUUGUUGUUGCUGCUUUUGC-GUUGCAGUUGC ..(((((......((((((((((....)))))......)))))((((((((.((((...(((..........)))...)))).)))))-))))))))... ( -31.20) >DroYak_CAF1 64084 88 - 1 UUGCUGCAUUGAAUGCGAGUCAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUGCAGUUGCGUGUUGUUGCU------------GUUGCAGUUGC ...(((((((...((((((((((....)))))......))))))))))))..((((.(((((..(((......))).------------.))))).)))) ( -30.40) >consensus UUGCUGCAUUGAAUGCGAGUCAGUAAGCUGACAAUUAUUUGCAGAUGCAGAUGCAGAUGCAGUUGCGUGUUGUUGCUGCUGCUUUUGC_GUUGCAGUUGC ..((((((((...(((..(((((....)))))....((((((....))))))))))))))))).....((..((((.((.((....)).)).))))..)) (-27.04 = -27.77 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:42 2006