
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,687,903 – 5,688,020 |
| Length | 117 |
| Max. P | 0.934996 |

| Location | 5,687,903 – 5,688,020 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
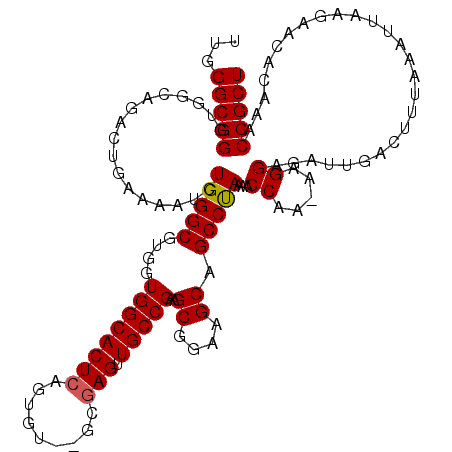
>3L_DroMel_CAF1 5687903 117 + 23771897 UUGGGCGGGUGGCAGACUGAAAAUGUGGGCGUGGUGGCACUCAGUGU--GCAAGUUGCCGAAGCGGAAGCAGCCUAAAACCAA-AAGGAGAUUGACUUUAAAUUGAGAACACAAACCGCU ...((((((((.(((.((.......(((((.((((.((((.....))--)).....))))..((....)).)))))...((..-..)))).))).(((......)))..)))...))))) ( -32.30) >DroSec_CAF1 60537 117 + 1 UUGGGCGGGUGGCAGACUGAAAACCUGGGCGUGGUGGCACUCAGUGU--GCGAGUUGCCGAAGCGGAAGCAGCCUAAAACCAA-AAGGAGAUUGACUUUAAAUUCAGAACACAAACCGCU ...((((((((.....(((((....(((((....((((((((.....--..))).)))))..((....)).)))))......(-(((........))))...)))))..)))...))))) ( -35.90) >DroSim_CAF1 60810 117 + 1 UUGGGCGGGUGGCAGACUGAAAACCUGGGCGUGGUGGCACUCAGUGU--GCGAGUUGCCGAGGCGGAAGCUGCCUAAAACCAA-AAGGAGAUUGACUUUAAAUUCAGAACACAAACCGCU ...(((((..(((((.((.....(((.((((.(.(.((((.....))--)).).))))).)))....)))))))........(-(((........))))................))))) ( -37.50) >DroEre_CAF1 64005 117 + 1 UUGGGCGGUGGGAAGCCUCAAAAUCUGGGCGUGGUGGCACUCAGUGU--GGGAGUUGCCGAAGCGGAAGCAGCCCAAAACCAA-AAGGAGAUUGACUUUAAAUUAAGAACACAUACCGCU ...((((((((....))....(((((((((.(((..((.(((.....--))).))..)))..((....)).))))....((..-..))))))).....................)))))) ( -39.60) >DroYak_CAF1 62603 120 + 1 UUGGGCGGUUGGCAGCCUCAAAAUCUGGGCGUGGUGGCACUCAGUGUGUGUGAGUUGCCGAAGCGGAAGCAGCCUAAAACCAAAAAGGAGAUUGACUUUAAAUUAAGAGCACAAACCGCU ...(((((((.((........(((((((((....(((((((((.......)))).)))))..((....)).))))....((.....)))))))..(((......))).))...))))))) ( -38.90) >consensus UUGGGCGGGUGGCAGACUGAAAAUCUGGGCGUGGUGGCACUCAGUGU__GCGAGUUGCCGAAGCGGAAGCAGCCUAAAACCAA_AAGGAGAUUGACUUUAAAUUAAGAACACAAACCGCU ...(((((.................(((((....((((((((.........))).)))))..((....)).)))))...((.....))...........................))))) (-29.44 = -29.48 + 0.04)


| Location | 5,687,903 – 5,688,020 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5687903 117 - 23771897 AGCGGUUUGUGUUCUCAAUUUAAAGUCAAUCUCCUU-UUGGUUUUAGGCUGCUUCCGCUUCGGCAACUUGC--ACACUGAGUGCCACCACGCCCACAUUUUCAGUCUGCCACCCGCCCAA .((((...(((.........((((..(((.......-)))..))))(((((.....((....)).....((--((.....)))).................)))))...))))))).... ( -25.90) >DroSec_CAF1 60537 117 - 1 AGCGGUUUGUGUUCUGAAUUUAAAGUCAAUCUCCUU-UUGGUUUUAGGCUGCUUCCGCUUCGGCAACUCGC--ACACUGAGUGCCACCACGCCCAGGUUUUCAGUCUGCCACCCGCCCAA (((((...(((..(((((...((((........)))-)....)))))..)))..)))))..(((.....((--(.((((((.(((..........))).)))))).))).....)))... ( -31.50) >DroSim_CAF1 60810 117 - 1 AGCGGUUUGUGUUCUGAAUUUAAAGUCAAUCUCCUU-UUGGUUUUAGGCAGCUUCCGCCUCGGCAACUCGC--ACACUGAGUGCCACCACGCCCAGGUUUUCAGUCUGCCACCCGCCCAA .((((...............((((..(((.......-)))..))))(((((((...((((.(((.....((--((.....))))......))).))))....)).)))))..)))).... ( -34.70) >DroEre_CAF1 64005 117 - 1 AGCGGUAUGUGUUCUUAAUUUAAAGUCAAUCUCCUU-UUGGUUUUGGGCUGCUUCCGCUUCGGCAACUCCC--ACACUGAGUGCCACCACGCCCAGAUUUUGAGGCUUCCCACCGCCCAA .(((((.............((((((((.....((..-..))...(((((.((....))...((((.(((..--.....))))))).....)))))))))))))((....))))))).... ( -33.70) >DroYak_CAF1 62603 120 - 1 AGCGGUUUGUGCUCUUAAUUUAAAGUCAAUCUCCUUUUUGGUUUUAGGCUGCUUCCGCUUCGGCAACUCACACACACUGAGUGCCACCACGCCCAGAUUUUGAGGCUGCCAACCGCCCAA .((((((.((((.(((((..((((..(((........)))..))))(((.((....))...((((.((((.......)))))))).....)))......))))))).)).)))))).... ( -35.40) >consensus AGCGGUUUGUGUUCUGAAUUUAAAGUCAAUCUCCUU_UUGGUUUUAGGCUGCUUCCGCUUCGGCAACUCGC__ACACUGAGUGCCACCACGCCCAGAUUUUCAGUCUGCCACCCGCCCAA .((((...(((..(((((..((((..(((........)))..))))(((.((....))...((((.((((.......)))))))).....)))......)))))..)))...)))).... (-27.86 = -27.70 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:38 2006