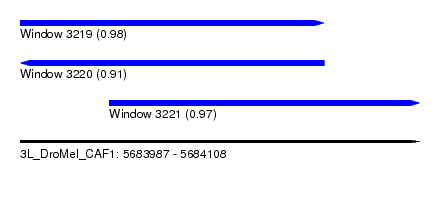
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,683,987 – 5,684,108 |
| Length | 121 |
| Max. P | 0.978284 |

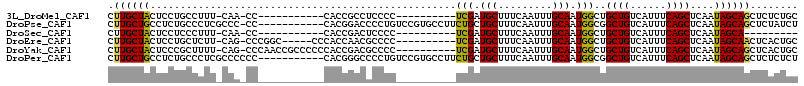
| Location | 5,683,987 – 5,684,079 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.41 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.05 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5683987 92 + 23771897 CUUGCUACUCCUGCCUUU-CAA-CC-----------CACCGCCUCCCC----------UCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAGCUCUCUGC .((((((.....(((.((-(((-..-----------....((.((...----------..)).)).......))).)).))).((((......))))....))))))........ ( -15.52) >DroPse_CAF1 60308 103 + 1 CUUGCUGCCUCUGCCCUCGCCC-CC-----------CACGGACCCCUGUCCGUGCCUUCUGCUGCUUUCAAUUUGCAAUGGCGGCUGUCAUUUCAGCUCAAUAGCAGCUCUAUCU ...(((((.........((((.-..-----------(((((((....)))))))........(((.........)))..))))((((......))))......)))))....... ( -32.20) >DroSec_CAF1 56529 83 + 1 CUUGCUACUCCUCCCUUU-CAA-CC-----------CACCGACUCCCC----------UCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCA--------- ..(((((...........-...-.(-----------((.(((......----------))).(((.........))).)))..((((......))))....)))))--------- ( -13.70) >DroEre_CAF1 60008 98 + 1 CUUGCUACUCCUGCUCUU-CAG-CCCGGC-----CCCACCAACGCCCC----------UCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAACUCACUGC .((((((...........-.((-((.(((-----.........)))..----------....(((.........)))..))))((((......))))....))))))........ ( -20.00) >DroYak_CAF1 58190 103 + 1 CUUGCUACUCCCGCUUUU-CAG-CCCAACCGCCCCCCACCGACGCCCC----------UCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAGCUCACUGC .((((((...........-.((-((..............(((......----------))).(((.........)))..))))((((......))))....))))))........ ( -18.40) >DroPer_CAF1 60864 104 + 1 CUUGCUGCCUCUGCCCUCGCCCCCC-----------CACGGGCCCCUGUCCGUGCCUUCUGCUGCUUUCAAUUUGCAAUGGCGGCUGUCAUUUCAGCUCAAUAGCAGCUCUCUCU ...(((((.........((((....-----------(((((((....)))))))........(((.........)))..))))((((......))))......)))))....... ( -31.60) >consensus CUUGCUACUCCUGCCCUU_CAA_CC___________CACCGACCCCCC__________UCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAGCUCUCUGC .((((((...................................................(((.(((.........))).)))..((((......))))....))))))........ (-12.66 = -12.05 + -0.61)


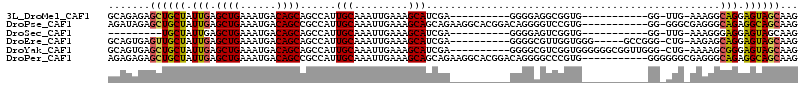
| Location | 5,683,987 – 5,684,079 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.41 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -14.93 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5683987 92 - 23771897 GCAGAGAGCUGCUAUUGAGCUGAAAUGACAGCAGCCAUUGCAAAUUGAAAGCAUCGA----------GGGGAGGCGGUG-----------GG-UUG-AAAGGCAGGAGUAGCAAG .......((((((.(((.((((......))))..(((((((...((((.....))))----------......))))))-----------).-...-.....))).))))))... ( -26.90) >DroPse_CAF1 60308 103 - 1 AGAUAGAGCUGCUAUUGAGCUGAAAUGACAGCCGCCAUUGCAAAUUGAAAGCAGCAGAAGGCACGGACAGGGGUCCGUG-----------GG-GGGCGAGGGCAGAGGCAGCAAG .......((((((.(((.((((......))))((((.((((.........)))).......(((((((....)))))))-----------..-.))))....))).))))))... ( -38.00) >DroSec_CAF1 56529 83 - 1 ---------UGCUAUUGAGCUGAAAUGACAGCAGCCAUUGCAAAUUGAAAGCAUCGA----------GGGGAGUCGGUG-----------GG-UUG-AAAGGGAGGAGUAGCAAG ---------(((((((..((((......))))..((.((.(...((.((..((((((----------......))))))-----------..-)).-)).).))))))))))).. ( -20.50) >DroEre_CAF1 60008 98 - 1 GCAGUGAGUUGCUAUUGAGCUGAAAUGACAGCAGCCAUUGCAAAUUGAAAGCAUCGA----------GGGGCGUUGGUGGG-----GCCGGG-CUG-AAGAGCAGGAGUAGCAAG ........((((((((..((((......)))).((((.(((...((((.....))))----------...))).))))...-----.((..(-((.-...))).)))))))))). ( -28.70) >DroYak_CAF1 58190 103 - 1 GCAGUGAGCUGCUAUUGAGCUGAAAUGACAGCAGCCAUUGCAAAUUGAAAGCAUCGA----------GGGGCGUCGGUGGGGGGCGGUUGGG-CUG-AAAAGCGGGAGUAGCAAG ((..(.(((((((.....((((......)))).(((..(((...((((.....))))----------...)))..)))....))))))).)(-((.-...))).......))... ( -30.20) >DroPer_CAF1 60864 104 - 1 AGAGAGAGCUGCUAUUGAGCUGAAAUGACAGCCGCCAUUGCAAAUUGAAAGCAGCAGAAGGCACGGACAGGGGCCCGUG-----------GGGGGGCGAGGGCAGAGGCAGCAAG .......((((((.(((.((((......))))((((.((((.........)))).......(((((........)))))-----------....))))....))).))))))... ( -33.50) >consensus GCAGAGAGCUGCUAUUGAGCUGAAAUGACAGCAGCCAUUGCAAAUUGAAAGCAUCGA__________GGGGAGUCGGUG___________GG_CUG_AAAGGCAGGAGUAGCAAG .......((((((.(((.((((......))))......(((.........))).................................................))).))))))... (-14.93 = -14.73 + -0.19)


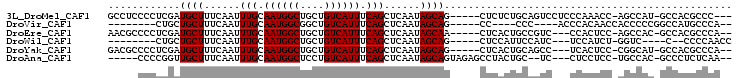
| Location | 5,684,014 – 5,684,108 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.47 |
| Mean single sequence MFE | -19.47 |
| Consensus MFE | -10.67 |
| Energy contribution | -10.67 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5684014 94 + 23771897 GCCUCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAG-----CUCUCUGCAGUCCUCCCAAACC-AGCCAU-GCCACGCCC--- ((.((.....)).)).........(((.(((((((((......))))......((((-----....))))..............-))))))-)).......--- ( -20.90) >DroVir_CAF1 70544 81 + 1 --------CUGCUGCUUUCAAUUUGCAAUGGCGGCUGUCAUUUCAGCUCAAUAGCAG-----CC----CCC----ACCCACAACCACCCCCGGCCAUGCCCA-- --------..((((((......(((.((((((....)))))).)))......)))))-----).----...----................(((...)))..-- ( -17.80) >DroEre_CAF1 60041 92 + 1 AACGCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAA-----CUCACUGCCGUC---CCACUCC-AGCCAC-GCCACGCCCA-- ...((....((.(((.........))).(((((((((......))))......(((.-----.....)))....---.......-))))))-)....))...-- ( -16.90) >DroWil_CAF1 72510 81 + 1 --------CUGCUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAG-----CUCCAUUCCAUC---UCCAUCU-GGUC----C--CCCCAACC --------..((((((......(((.((((((....)))))).)))......)))))-----)...........---......(-((..----.--..)))... ( -18.20) >DroYak_CAF1 58228 92 + 1 GACGCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAG-----CUCACUGCAGCC---UCACUCC-CGGCAU-GCCACGCCCA-- ((.((.....((.....))....((((.((((((((((............)))))))-----).)).)))))).---)).....-.(((..-.....)))..-- ( -23.10) >DroAna_CAF1 58017 90 + 1 -----CCCCGGUUGCUUUCAAUUUGCAAUGGCUCCUGUCAUUUCAGCUCAAUAGCAGUAGAGCCUACUGC--UC---CUCCUCC-UGCCAC-GCCCUCUCAA-- -----....((((((.........))).((((..(((......)))......((((((((...)))))))--).---.......-.)))).-))).......-- ( -19.90) >consensus _____CCCCCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAG_____CUCACUGCCGUC___CCACUCC_AGCCAC_GCCACGCCCA__ ............((((......(((.((((((....)))))).)))......))))................................................ (-10.67 = -10.67 + -0.00)
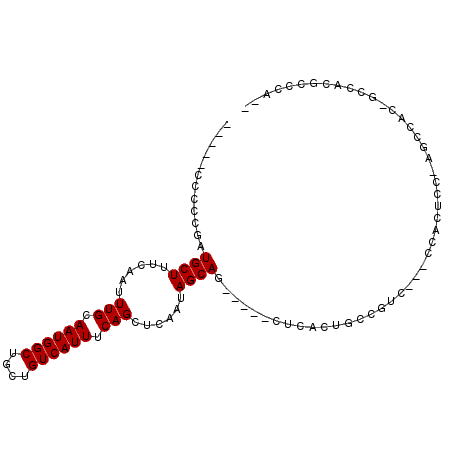
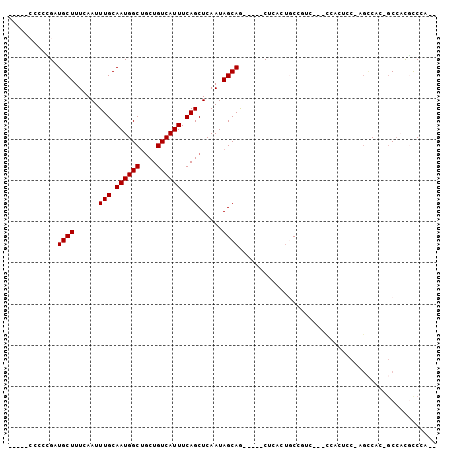
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:35 2006