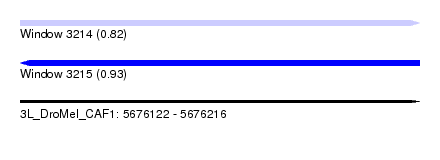
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,676,122 – 5,676,216 |
| Length | 94 |
| Max. P | 0.930433 |

| Location | 5,676,122 – 5,676,216 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.79 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5676122 94 + 23771897 ---AGUUCCACUCGACAGUUUCCGCUCAGCU--UGAUGAU-UUCAUGGAAAACUUCAUUGAGCA-AGCGGAAACUAUUUCGUUUUGGCGCGGAGUG--------CAGGU------- ---.....(((((...((((((((((..(((--..(((((-(((...))))...))))..))).-))))))))))....(((......))))))))--------.....------- ( -33.20) >DroVir_CAF1 61704 106 + 1 ---AGUUCCGCUCGACAGCUUCCGUUCGGCU--UGAUGAU-UUCAUGGAAAACUUCAUUGAGCA-AGCGGAAACUAUUUCAUUU-GGC--GCUAUAAAUGUUGUCAGGUGUACUUU ---(((..(((..(((((((((((((..(((--..(((((-(((...))))...))))..))).-)))))))........((((-(..--....)))))))))))..))).))).. ( -31.10) >DroPse_CAF1 50371 98 + 1 AACAGUUCCGCUCGACAGCUUCCGCUUGGCUGAUGAUGAUUUUCAUGGAAAACUUCAUUGAGCA-AGCGGAAACUAUUUCAUUUUGGU--GGAGUG--------CCAAU------- ........(((((.((((.(((((((((.(((((((.(.((((....)))).).))))).))))-))))))).))....(.....)))--.)))))--------.....------- ( -28.80) >DroGri_CAF1 53532 109 + 1 ACCAGUUCCGCUCGACAGCUUCCGUUCGGCU--UGAUGAU-UUCAUGAAAAACUUCAUUGAGCA-AGCGGAAACUAUUUCAUUU-GGC--GCAAUAAAUGUUGUCAGUUGAAUGUU ....((((.(((.(((((((((((((..(((--..((((.-.............))))..))).-)))))))........((((-(..--....)))))))))))))).))))... ( -31.64) >DroWil_CAF1 64342 93 + 1 AACAGUUCCGCUCGACAGCUUCCGCUUGGCU--UGAUGAU-UUCAUGGAAAACUUCAUUGAGCA-AGCGGAAACUAUUUCAUUUGGUC--AGAG----------CAGGA------- ......((((((((((((.(((((((((.((--..(((((-(((...))))...))))..))))-))))))).))....(....))))--.)))----------).)))------- ( -33.30) >DroAna_CAF1 50140 98 + 1 AACAGUUCCGCUCGACAGCUUCCGCUCGGCU--UGAUGAU-UUCAUGGAAAACUUCAUUGAGUAAAGCGGAAACUAUUUCAUUUUGGU--GGAGUG-------CCAGGU------U (((.....(((((.((((.(((((((..(((--..(((((-(((...))))...))))..)))..))))))).))....(.....)))--.)))))-------....))------) ( -29.20) >consensus AACAGUUCCGCUCGACAGCUUCCGCUCGGCU__UGAUGAU_UUCAUGGAAAACUUCAUUGAGCA_AGCGGAAACUAUUUCAUUUUGGC__GGAGUG________CAGGU_______ .....(((((((....)))((((((...(((...(((((...............))))).)))...))))))..................))))...................... (-15.98 = -15.79 + -0.19)

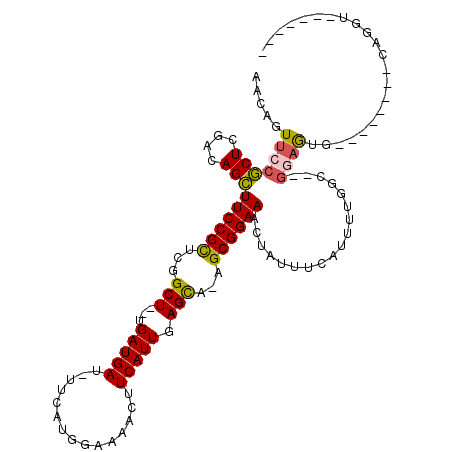
| Location | 5,676,122 – 5,676,216 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5676122 94 - 23771897 -------ACCUG--------CACUCCGCGCCAAAACGAAAUAGUUUCCGCU-UGCUCAAUGAAGUUUUCCAUGAA-AUCAUCA--AGCUGAGCGGAAACUGUCGAGUGGAACU--- -------.....--------(((((..((......))..((((((((((((-((((..((((..((......)).-.))))..--))).))))))))))))).))))).....--- ( -31.50) >DroVir_CAF1 61704 106 - 1 AAAGUACACCUGACAACAUUUAUAGC--GCC-AAAUGAAAUAGUUUCCGCU-UGCUCAAUGAAGUUUUCCAUGAA-AUCAUCA--AGCCGAACGGAAGCUGUCGAGCGGAACU--- ..(((.(..(((((..(((((.....--...-)))))....((((((((.(-((((..((((..((......)).-.))))..--)).))).))))))))))).))..).)))--- ( -23.80) >DroPse_CAF1 50371 98 - 1 -------AUUGG--------CACUCC--ACCAAAAUGAAAUAGUUUCCGCU-UGCUCAAUGAAGUUUUCCAUGAAAAUCAUCAUCAGCCAAGCGGAAGCUGUCGAGCGGAACUGUU -------.((((--------......--.)))).....(((((((.(((((-((((..((((.((((((...))))))..)))).)))..(((....)))...))))))))))))) ( -28.40) >DroGri_CAF1 53532 109 - 1 AACAUUCAACUGACAACAUUUAUUGC--GCC-AAAUGAAAUAGUUUCCGCU-UGCUCAAUGAAGUUUUUCAUGAA-AUCAUCA--AGCCGAACGGAAGCUGUCGAGCGGAACUGGU ..(((((..(((((....((((((..--...-.))))))..((((((((.(-((((..((((..((......)).-.))))..--)).))).))))))))))).))..))).)).. ( -25.90) >DroWil_CAF1 64342 93 - 1 -------UCCUG----------CUCU--GACCAAAUGAAAUAGUUUCCGCU-UGCUCAAUGAAGUUUUCCAUGAA-AUCAUCA--AGCCAAGCGGAAGCUGUCGAGCGGAACUGUU -------..(((----------(((.--(((..........((((((((((-((((..((((..((......)).-.))))..--)).)))))))))))))))))))))....... ( -33.90) >DroAna_CAF1 50140 98 - 1 A------ACCUGG-------CACUCC--ACCAAAAUGAAAUAGUUUCCGCUUUACUCAAUGAAGUUUUCCAUGAA-AUCAUCA--AGCCGAGCGGAAGCUGUCGAGCGGAACUGUU .------.(((((-------......--.))).......(((((((((((((..((..((((..((......)).-.))))..--))..))))))))))))).....))....... ( -25.20) >consensus _______ACCUG________CACUCC__GCCAAAAUGAAAUAGUUUCCGCU_UGCUCAAUGAAGUUUUCCAUGAA_AUCAUCA__AGCCGAGCGGAAGCUGUCGAGCGGAACUGUU .......................................((((((((((((..(((...(((.(..(((...)))...).)))..)))..)))))))))))).............. (-17.96 = -18.18 + 0.22)
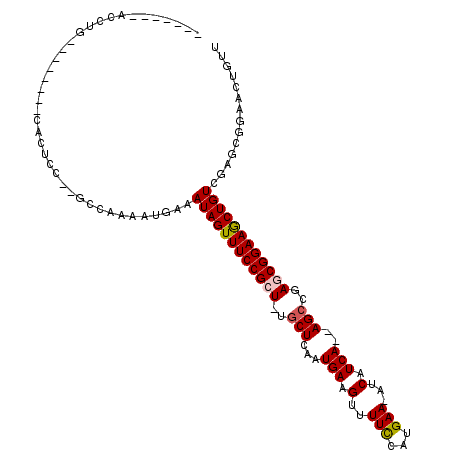
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:30 2006