| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,664,230 – 5,664,331 |
| Length | 101 |
| Max. P | 0.828972 |

| Location | 5,664,230 – 5,664,331 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.66 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5664230 101 + 23771897 UAGAGCCAAAUCCUGAGUGCACACAGCAGGAGCAACUAU--------CCAGCCAGCUGGAGCGGCAUCAGAAGCAAAUGCACUCGGAAAAAUAAGACAGUACUUGCU----GA ............(((((((((....((..((((.....(--------((((....)))))...)).))....))...)))))))))..........((((....)))----). ( -29.50) >DroSec_CAF1 36882 101 + 1 UAGAGCCAAAUCCUGAGUGCACACAGCAGGAGCAGCUAU--------CCAGCCAGCUGGAGCGGCAUCAGAAGCAAAUGCACUCGGAGAAAUAGGACUGUACUUGCU----AA ..(((((...(((((..((....)).)))))...((..(--------((((....)))))))))).))...(((((.((((.((..(....)..)).)))).)))))----.. ( -32.30) >DroSim_CAF1 37970 100 + 1 UAGAGCCAAAU-CUGAGUGCACACAGCAGGAGCAGCUAU--------CCAGCCAGCUGGAGCGGCAUCAGAAGCAAAUGCACUCGGAGAAAUAGGACAGUACUCGAU----AA ..(((((...(-(((((((((....((....)).(((.(--------((((....)))))..)))............))))))))))......))......)))...----.. ( -30.90) >DroEre_CAF1 40136 113 + 1 CAGAGCAAAAUCCUGAGUGCACACAGCAGGAGCAGCCAUCCAGCCAUCCAGCCAGCUGGAGCGGCAUCAGAAGCAAGUGCACUUGGAGAAAUAAGGCAGUACUUGCUGGCUAA ....((....(((((..((....)).)))))))((((.....(((.(((((....)))))..)))......(((((((((.((((......))))...))))))))))))).. ( -42.10) >DroYak_CAF1 37647 105 + 1 CAGAGCAAAAUCCUGAGUGCACACAGCAGGAGCAGCCAA--------CCAGCCAGCUGGAGCGGCAUUAGAAGCAAGUGCACUCGGAGAAAUAAGGCAGUACUUGCUGGCUAA ..........((((((((((((...((....)).(((..--------((((....))))...)))...........)))))))))).))....((.((((....)))).)).. ( -37.90) >consensus UAGAGCCAAAUCCUGAGUGCACACAGCAGGAGCAGCUAU________CCAGCCAGCUGGAGCGGCAUCAGAAGCAAAUGCACUCGGAGAAAUAAGACAGUACUUGCU____AA ...(((......(((((((((....((.((.....))..........((((....)))).)).((.......))...)))))))))......(((......))))))...... (-22.90 = -22.66 + -0.24)

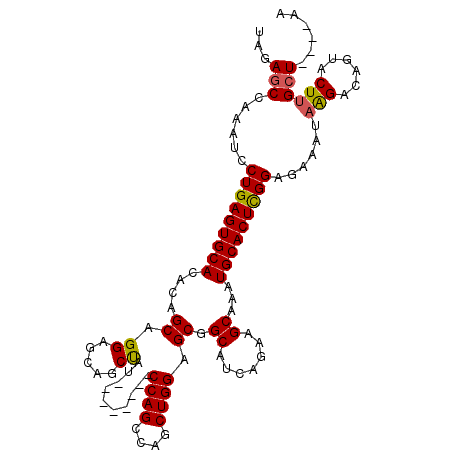

| Location | 5,664,230 – 5,664,331 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -33.49 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5664230 101 - 23771897 UC----AGCAAGUACUGUCUUAUUUUUCCGAGUGCAUUUGCUUCUGAUGCCGCUCCAGCUGGCUGG--------AUAGUUGCUCCUGCUGUGUGCACUCAGGAUUUGGCUCUA ..----((((((......))).....(((((((((((..((....((.((.(((((((....))))--------)..)).))))..))...)))))))).)))....)))... ( -30.80) >DroSec_CAF1 36882 101 - 1 UU----AGCAAGUACAGUCCUAUUUCUCCGAGUGCAUUUGCUUCUGAUGCCGCUCCAGCUGGCUGG--------AUAGCUGCUCCUGCUGUGUGCACUCAGGAUUUGGCUCUA ..----.........((..(((..(((..((((((((..((....((.((.(((((((....))))--------)..)).))))..))...)))))))).)))..)))..)). ( -32.60) >DroSim_CAF1 37970 100 - 1 UU----AUCGAGUACUGUCCUAUUUCUCCGAGUGCAUUUGCUUCUGAUGCCGCUCCAGCUGGCUGG--------AUAGCUGCUCCUGCUGUGUGCACUCAG-AUUUGGCUCUA ..----...(((.......(((..(((..((((((((..((....((.((.(((((((....))))--------)..)).))))..))...))))))))))-)..)))))).. ( -30.81) >DroEre_CAF1 40136 113 - 1 UUAGCCAGCAAGUACUGCCUUAUUUCUCCAAGUGCACUUGCUUCUGAUGCCGCUCCAGCUGGCUGGAUGGCUGGAUGGCUGCUCCUGCUGUGUGCACUCAGGAUUUUGCUCUG .((((((((((((...(((((........))).)))))))).(((...((((.(((((....))))))))).))))))))).(((((..((....)).))))).......... ( -38.00) >DroYak_CAF1 37647 105 - 1 UUAGCCAGCAAGUACUGCCUUAUUUCUCCGAGUGCACUUGCUUCUAAUGCCGCUCCAGCUGGCUGG--------UUGGCUGCUCCUGCUGUGUGCACUCAGGAUUUUGCUCUG ..(((..(((.....)))........(((((((((((..((.......((((..((((....))))--------.)))).......))...)))))))).)))....)))... ( -35.24) >consensus UU____AGCAAGUACUGUCUUAUUUCUCCGAGUGCAUUUGCUUCUGAUGCCGCUCCAGCUGGCUGG________AUAGCUGCUCCUGCUGUGUGCACUCAGGAUUUGGCUCUA ......((((((((......))))).(((((((((((..((....((.((.(((((((....))))..........))).))))..))...)))))))).)))....)))... (-25.54 = -25.54 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:23 2006