
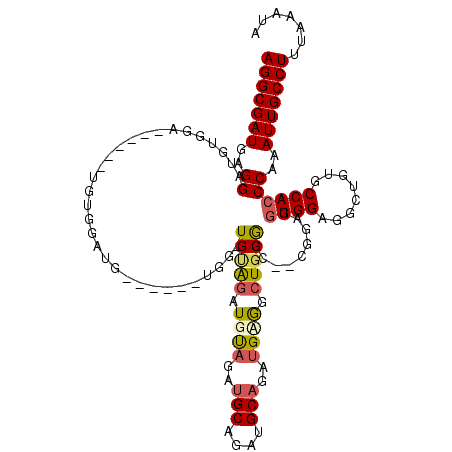
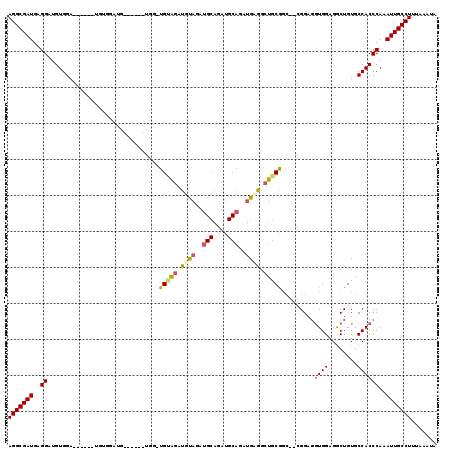
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,663,124 – 5,663,229 |
| Length | 105 |
| Max. P | 0.992358 |

| Location | 5,663,124 – 5,663,229 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.08 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -14.58 |
| Energy contribution | -15.06 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5663124 105 + 23771897 UAUUUAAAGGCAAUUUGGGUGGCACAGCCUCCACAUCCG--GACGCAGCCUCAUCUGCAUCUGCAUCUACAUCUACAUCCA------CAUCCACA------UCCACAUCCUCAUCGCCU .......((((.((..(((((.....((.(((......)--)).)).........(((....)))................------........------....)))))..)).)))) ( -20.80) >DroSec_CAF1 35805 93 + 1 UAUUUAAAGGCAAUUUGGGUGGCACAGUCCCCACCUCCG--GCCGCAGCCACAUCUGCAUCUGCAUCUACAUCCUCA------------------------UCCACAUCCUCAUCGCCU .......((((.....((((((........))))))..(--((....))).....(((....)))............------------------------..............)))) ( -19.50) >DroSim_CAF1 36884 93 + 1 UAUUUAAAGGCAAUUUGGGUGGCACAGCCUCCACCUCCG--GCCGCAGCCACAUCUGCAUCUGCAUCUGCAUCUACA------------------------UCCACAUCCUCAUCGCCU .......((((.((..(((((((.(.(((.........)--)).)..)))))...((((........))))......------------------------.......))..)).)))) ( -21.90) >DroEre_CAF1 39059 111 + 1 UAUUUAAAGGCAAUUUGGGUGGCACAGCCUCCAUCUCCU--GCCGCAGCCUCAUCUGCAUCUGCAUUUACAUCCACAUCCACUUUCACAUCCACA------UCCACAUCCUCAUCGCCU .......((((.((..((((((((..............)--)))((((......)))).....................................------.....))))..)).)))) ( -18.04) >DroYak_CAF1 36526 107 + 1 UAUUUAAAGGCAAUUUGGGUGGCACAGCCUCCACCGCCUCAUCUGCAUCUGC------------AUCUACAUCUGCAGCCACAUUCACAUCCACAUCCACUUCCACAUCCUCAUCGCCU .......((((......(((((........))))).......(((((..((.------------.....))..))))).....................................)))) ( -19.30) >consensus UAUUUAAAGGCAAUUUGGGUGGCACAGCCUCCACCUCCG__GCCGCAGCCACAUCUGCAUCUGCAUCUACAUCUACA_CCA______CAUCCACA______UCCACAUCCUCAUCGCCU .......((((....((((.(((...)))))))...........((((......)))).........................................................)))) (-14.58 = -15.06 + 0.48)

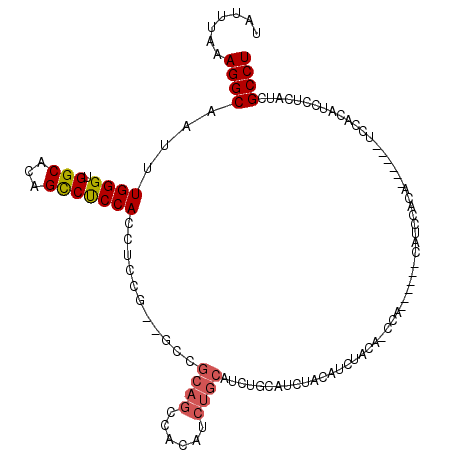
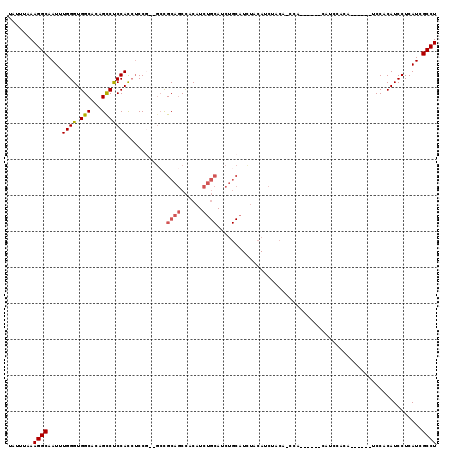
| Location | 5,663,124 – 5,663,229 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.08 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -20.62 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5663124 105 - 23771897 AGGCGAUGAGGAUGUGGA------UGUGGAUG------UGGAUGUAGAUGUAGAUGCAGAUGCAGAUGAGGCUGCGUC--CGGAUGUGGAGGCUGUGCCACCCAAAUUGCCUUUAAAUA (((((((..((((.((((------((..(...------((..((((..(((....)))..))))..))...)..))))--)).))((((........))))))..)))))))....... ( -35.50) >DroSec_CAF1 35805 93 - 1 AGGCGAUGAGGAUGUGGA------------------------UGAGGAUGUAGAUGCAGAUGCAGAUGUGGCUGCGGC--CGGAGGUGGGGACUGUGCCACCCAAAUUGCCUUUAAAUA (((((((..((.......------------------------............(((....)))...(((((.(((((--(........)).)))))))))))..)))))))....... ( -27.90) >DroSim_CAF1 36884 93 - 1 AGGCGAUGAGGAUGUGGA------------------------UGUAGAUGCAGAUGCAGAUGCAGAUGUGGCUGCGGC--CGGAGGUGGAGGCUGUGCCACCCAAAUUGCCUUUAAAUA (((((((..((.......------------------------((((..(((....)))..))))...(((((.(((((--(.........)))))))))))))..)))))))....... ( -36.80) >DroEre_CAF1 39059 111 - 1 AGGCGAUGAGGAUGUGGA------UGUGGAUGUGAAAGUGGAUGUGGAUGUAAAUGCAGAUGCAGAUGAGGCUGCGGC--AGGAGAUGGAGGCUGUGCCACCCAAAUUGCCUUUAAAUA (((((((..((......(------(.((.((.((....((.((....)).))....)).)).)).))..(((.(((((--...........))))))))..))..)))))))....... ( -27.30) >DroYak_CAF1 36526 107 - 1 AGGCGAUGAGGAUGUGGAAGUGGAUGUGGAUGUGAAUGUGGCUGCAGAUGUAGAU------------GCAGAUGCAGAUGAGGCGGUGGAGGCUGUGCCACCCAAAUUGCCUUUAAAUA (((((((..((..((((........................(((((..(((....------------)))..))))).....(((((....))))).))))))..)))))))....... ( -30.00) >consensus AGGCGAUGAGGAUGUGGA______UGUGGAUG______UGG_UGUAGAUGUAGAUGCAGAUGCAGAUGAGGCUGCGGC__CGGAGGUGGAGGCUGUGCCACCCAAAUUGCCUUUAAAUA (((((((..((...............................(((((.((((..(((....)))..)))).))))).........((((........))))))..)))))))....... (-20.62 = -21.20 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:19 2006