
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,639,902 – 5,640,009 |
| Length | 107 |
| Max. P | 0.550143 |

| Location | 5,639,902 – 5,640,009 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -24.31 |
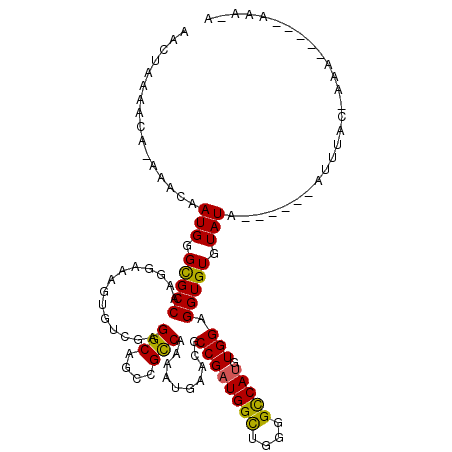
| Consensus MFE | -19.21 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
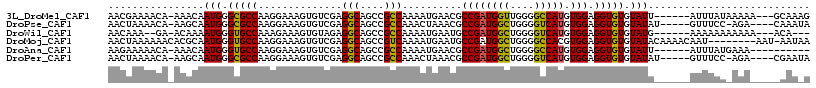
>3L_DroMel_CAF1 5639902 107 + 23771897 AACGAAAACA-AAACAAUGGGCGCCAAGGAAAGUGUCGAGGCAGCCGCCAAAAUGAACGCCGAUGGUUGGGGCCAUGUGGAGGUGUGUAUU------AUUUAUAAAAA---GCAAAG ..........-(((.((((.(((((..............(((....)))..........((((((((....))))).))).))))).))))------.))).......---...... ( -23.00) >DroPse_CAF1 10718 106 + 1 AACUAAAACA-AAGCAAUGGGCGCCAAGGAAAGUGUCGAGGCAGCCGCCAAACUAAACGCCGAUGGCUGGGGUCAUGUGGAGGUGUGUAUAU-----GUUUCC-AGA----CAAAUA ........(.-.(((.((.((((...((((.....))..(((....)))...))...)))).)).)))..)(((...((((((..(....).-----.)))))-)))----)..... ( -25.50) >DroWil_CAF1 12564 102 + 1 AACAAA--GA-ACAAAAUGGGUGCCAAAGAAAGUGUAGAGGCAGCCGCCAAAAUGAAUGCCGAUGGCUGGGGUCAUGUGGAGGUGUGUAUG------AAAAAAAAAAA---ACA--- .(((..--..-((.....((.((((..............)))).)).(((..((((..(((...))).....)))).)))..)).)))...------...........---...--- ( -18.54) >DroMoj_CAF1 16418 108 + 1 AACUAAAAAACACGCAAUGGGUGCCAAGGAAAGUGUCGAGGCAGCCGUCAAAAUGAAUGCCGAUGGCUGGGGCCACGUGGAGGUGUGUAUACAAAACAAU--------AAU-AAUAA .........((((((.((((((.(((.......(((....)))((((((............))))))))).))).)))....))))))............--------...-..... ( -28.70) >DroAna_CAF1 10215 100 + 1 AAGAAAAACA-AAACAAUGGGUGCCAAGGAAAGUGUCGAGGCAGCCGCCAAAAUGAACGCCGAUGGCUGGGGCCAUGUGGAGGUGUGUAUU------AUUUAUGAAA---------- ..........-.......((.((((..((......))..)))).)).....((((.(((((.(((((....))))).....))))).))))------..........---------- ( -24.60) >DroPer_CAF1 10845 106 + 1 AACUAAAACA-AAGCAAUGGGCGCCAAGGAAAGUGUCGAGGCAGCCGCCAAACUAAACGCCGAUGGCUGGGGUCAUGUGGAGGUGUGUAUAU-----GUUUCC-AGA----CGAAUA ........(.-.(((.((.((((...((((.....))..(((....)))...))...)))).)).)))..)(((...((((((..(....).-----.)))))-)))----)..... ( -25.50) >consensus AACUAAAACA_AAACAAUGGGCGCCAAGGAAAGUGUCGAGGCAGCCGCCAAAAUGAACGCCGAUGGCUGGGGCCAUGUGGAGGUGUGUAUA______AUUUAC_AAA_____AAA_A ................(((.(((((..............(((....)))..........((((((((....))))).))).))))).)))........................... (-19.21 = -18.60 + -0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:14 2006