
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,639,439 – 5,639,568 |
| Length | 129 |
| Max. P | 0.968987 |

| Location | 5,639,439 – 5,639,537 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -12.06 |
| Energy contribution | -13.40 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
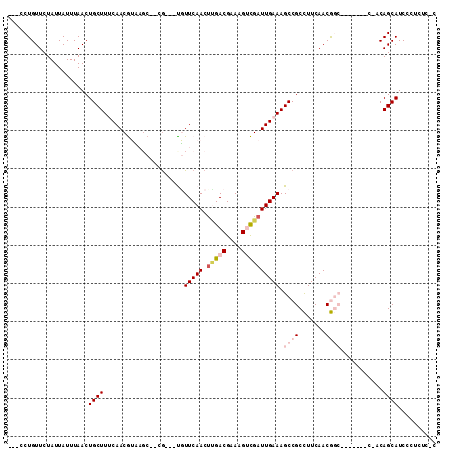
>3L_DroMel_CAF1 5639439 98 + 23771897 ---CCUGUUCUAUUAUUUAACUGCUUUCAACGUCAGCACCA---CGUUCAACCCGACGAAAGUCGAUUGAAAGCCGCCUUCAACGGC-------C-ACAGCAUCCCUCUCUC ---.((((..............(((((((((((.......)---)))......((((....))))..))))))).(((......)))-------.-))))............ ( -23.60) >DroVir_CAF1 17303 90 + 1 CUGCCUGUUCUAUUAUUUAACUGCUUACAACAGA-----CG---UUUUCAACUUGACGAAAGCUGAUUGAAAGCCGCUUUCAACA-----------ACAGCAUCCAUCG--- ....(((((.((.............)).))))).-----((---((........))))...((((.(((((((...)))))))..-----------.))))........--- ( -15.02) >DroPse_CAF1 10210 103 + 1 ---CCUGUUCUAUUAUUUAACUGCUUUCAACGUUAGC---G---UGUUCAACUUGACGAAAGUCGAUUGAAAGCCGCCUUCAACGGCAUCCAGCCACCAGCAUCCCUUUCUC ---.(((...............((((((((((....)---)---).......(((((....))))).))))))).(((......)))...)))................... ( -23.20) >DroMoj_CAF1 15850 90 + 1 CUGCCUGUUCUAUUAUUUAACUGCUUACAACAGA-----CG---UUUUCAACUGGACGAAAGCUGAUUGAAAGCCGCUUUCAACA-----------GCAGCAUCCGUCG--- ((((.((((...........(((.......))).-----.(---(((((((..((.(....)))..)))))))).......))))-----------)))).........--- ( -19.50) >DroAna_CAF1 9788 99 + 1 ---CCUGUUCUAUUAUUUAACUGCUUUCAACGUCAGCACAGCGCCGUUCAACUUGACGAAAGUCGAUUGAAAGCCGCCUUCAACGGC-------C--CAGCAUCCCUUUG-C ---...................((((((((.....((.....))........(((((....))))))))))))).(((......)))-------.--..(((......))-) ( -21.80) >DroPer_CAF1 10335 103 + 1 ---CCUGUUCUAUUAUUUAACUGCUUUCAACGUUAGC---G---UGUUCAACUUGACGAAAGUCGAUUGAAAGCCGCCUUCAACGGCAUCCAGCCACCAGCAUCCCUUUCUC ---.(((...............((((((((((....)---)---).......(((((....))))).))))))).(((......)))...)))................... ( -23.20) >consensus ___CCUGUUCUAUUAUUUAACUGCUUUCAACGUAAGC__CG___UGUUCAACUUGACGAAAGUCGAUUGAAAGCCGCCUUCAACGGC_______C_ACAGCAUCCCUCUC_C .....................((((.....................(((((.(((((....)))))))))).((((.......))))...........)))).......... (-12.06 = -13.40 + 1.34)


| Location | 5,639,474 – 5,639,568 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.77 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
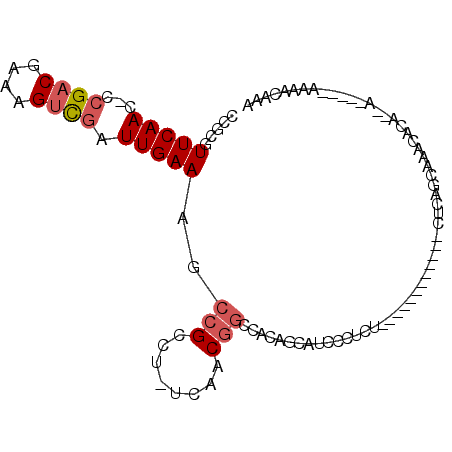
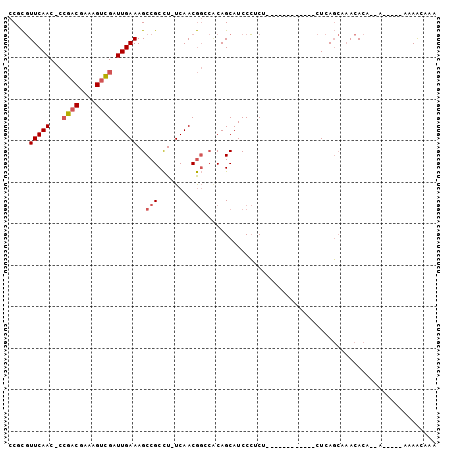
>3L_DroMel_CAF1 5639474 94 + 23771897 CCACGUUCAAC-CCGACGAAAGUCGAUUGAAAGCCGCCU-UCAACGGCCACAGCAUCCCUCUCUCUCUCUCUCUCUCAGCAAACACA--A-----AAAACAAA .....(((((.-.((((....)))).))))).((((...-....)))).......................................--.-----........ ( -16.20) >DroSec_CAF1 12478 85 + 1 CCGCGUUCAACCCCGACGAAAGUCGAUUGAAAGCCGCCU-UCAACGGCCACAGCAUCCCUCU----------CUCUCAGCAAACACA--A-----AAAACAAA ..((.(((((...((((....)))).))))).((((...-....))))....))........----------...............--.-----........ ( -18.00) >DroSim_CAF1 13089 84 + 1 CCGCGUUCAAC-CCGACGAAAGUCGAUUGAAAGCCGCCU-UCAACGGCCACAGCAUCCCUCU----------CUCUCAGCAAACACA--A-----AAAACAAA ..((.(((((.-.((((....)))).))))).((((...-....))))....))........----------...............--.-----........ ( -18.90) >DroEre_CAF1 12659 81 + 1 CCUCGUUCAAC-CCGACGAAAGUCGAUUGAAAGCCGCCU-UCAACGGACACAGCAUCCCUCU------------CUCAGCAAACACA--C------AAAAUAA ....((((...-.((((....)))).(((((.......)-)))).))))...((........------------....)).......--.------....... ( -13.70) >DroYak_CAF1 12407 88 + 1 GCACGUUCAAC-CCGACGAAAGUCGAUUGAAAGCCGCCUUUCAACGGACCCAGCAUCCCUCU------------CUCAGCACACACA--UCACAAAAAAGUAA ((..((((...-.((((....)))).(((((((....))))))).))))...))........------------.............--.............. ( -17.90) >DroMoj_CAF1 15884 81 + 1 -CGUUUUCAAC-UGGACGAAAGCUGAUUGAAAGCCGCUU-UCAACA---GCAGCAUCCGUCG------------UUCAC-UAAUACUCGACG---CGAGGGAG -(((((.....-.)))))...((((.(((((((...)))-))))))---))....(((.(((------------(((..-........)).)---))).))). ( -23.20) >consensus CCGCGUUCAAC_CCGACGAAAGUCGAUUGAAAGCCGCCU_UCAACGGCCACAGCAUCCCUCU____________CUCAGCAAACACA__A_____AAAACAAA .....(((((...((((....)))).)))))..(((........)))........................................................ ( -9.24 = -9.77 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:13 2006