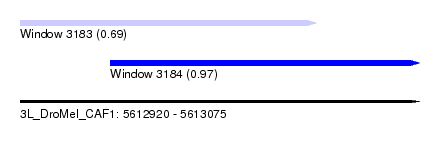
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,612,920 – 5,613,075 |
| Length | 155 |
| Max. P | 0.969913 |

| Location | 5,612,920 – 5,613,035 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5612920 115 + 23771897 AUAAAAAAAUCCUAUUGAUUUAUUGCCAUUAAAAGCAACAGCUGCUAAAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGCAGAGCAGCAACUGACCAAGUUAUAC ......(((((.....))))).((((........))))..((((((...((((((..........((.....))..........))))))..))))))((((.....)))).... ( -18.75) >DroSec_CAF1 31991 115 + 1 AUAAAAAAAUCCUAUUGAUUUAUUGCCAUUAAAAGCAACAGCUGCUAGAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGUAGAGCAGCAACAAACCANNNNNNNN ......(((((.....))))).((((........))))..((((((..(((((......)))))...............((....)).....))))))................. ( -16.50) >DroSim_CAF1 31372 114 + 1 AUCAA-AAAUCCUAUUGAUUUAUUGCCAUUAAAAGCAACAGCUGCUAGAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGUAGAGCAGCAACAAACCAAGUUAUAC .....-.........((((((.((((........))))..((((((..(((((......)))))...............((....)).....))))))........))))))... ( -16.90) >DroEre_CAF1 33446 114 + 1 AUAAA-AAAUCCUAUUGAUUUAUUGCCGCUAACAGCAAAAGCUGCUAGAGUUGCAAAAACAAUUAGUUAAGCGCCAAGCGACAAUUCAGUAGAGCAGCAACAGGCCAAGUUAUAU .....-.........((((((...(((((.....))....((((((.(((((((...(((.....)))..((.....))).)))))).....))))))....))).))))))... ( -24.80) >DroYak_CAF1 31905 113 + 1 AUAAA-AAAUCCUAUUGAUUUAUUGCCAUUAACAGCAACAGCUGCUAGAGUUGCAA-AACAAUUAGUAAAACACCUAAAGAUAAUUCAGUAGAGCAACAACAAACCCAGUUAUAC .....-.....(((((((.(((((((.(((..((((....))))....))).))..-.....((((........)))).))))).)))))))....................... ( -17.80) >consensus AUAAA_AAAUCCUAUUGAUUUAUUGCCAUUAAAAGCAACAGCUGCUAGAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGUAGAGCAGCAACAAACCAAGUUAUAC ......(((((.....))))).((((........))))..((((((..(((((......)))))...............((....)).....))))))................. (-14.38 = -14.58 + 0.20)

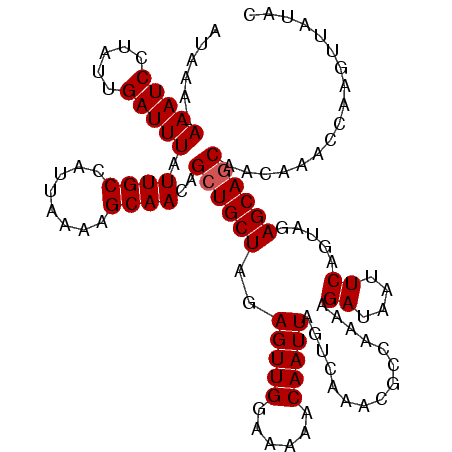
| Location | 5,612,955 – 5,613,075 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.55 |
| Mean single sequence MFE | -16.38 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5612955 120 + 23771897 CAACAGCUGCUAAAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGCAGAGCAGCAACUGACCAAGUUAUACAACAACAAUUCCCAUUCGCUUUUUAUUGUUCUUAACAAAA .....((((((...((((((..........((.....))..........))))))..))))))..........((((...(((((.((..............)).)))))..)))).... ( -16.99) >DroSec_CAF1 32026 103 + 1 CAACAGCUGCUAGAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGUAGAGCAGCAACAAACCANNNNNNNNNNNNNNNNNNNNNNNNNNN-----------------NNNN .....((((((..(((((......)))))...............((....)).....))))))....................................-----------------.... ( -13.40) >DroSim_CAF1 31406 103 + 1 CAACAGCUGCUAGAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGUAGAGCAGCAACAAACCAAGUUAUACAACAACAAUUCCCAUUUGC-----------------AAAA .....((((((..(((((......)))))...............((....)).....))))))....................................-----------------.... ( -13.40) >DroEre_CAF1 33480 118 + 1 CAAAAGCUGCUAGAGUUGCAAAAACAAUUAGUUAAGCGCCAAGCGACAAUUCAGUAGAGCAGCAACAGGCCAAGUUAUAUAACAA-AUUUCCUAUUUG-UAUAUAUUGUUCUAAACAAAA .....((((((.(((((((...(((.....)))..((.....))).)))))).....))))))...((..(((..((((((.(((-((.....)))))-)))))))))..))........ ( -25.20) >DroYak_CAF1 31939 113 + 1 CAACAGCUGCUAGAGUUGCAA-AACAAUUAGUAAAACACCUAAAGAUAAUUCAGUAGAGCAACAACAAACCCAGUUAUACAACCA-AAUCACUAUUAG-UUAUUAUUCUUCUU----AAA ....(((((.(((.(((((..-.((.....)).......(((..((....))..))).)))))(((.......))).........-.....))).)))-))............----... ( -12.90) >consensus CAACAGCUGCUAGAGUUGGAAAAACAAUUAGUCAAACGCCAAAAGAUAAUUCAGUAGAGCAGCAACAAACCAAGUUAUACAACAA_AAUUCCCAUUUG_U___UAUU_UUCU____AAAA .....((((((..(((((......)))))...............((....)).....))))))......................................................... (-11.40 = -11.60 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:00 2006