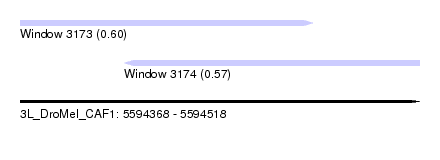
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,594,368 – 5,594,518 |
| Length | 150 |
| Max. P | 0.604956 |

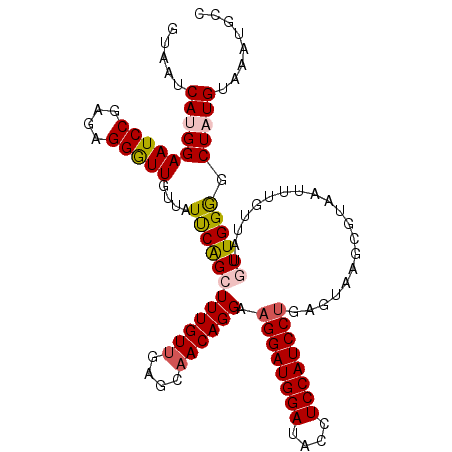
| Location | 5,594,368 – 5,594,478 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.02 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -11.34 |
| Energy contribution | -13.37 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5594368 110 + 23771897 CCU-UAAGGGUAACAUCAUUGCUUAUCAAACAGAUGUCCUGGCAUUUACAUAGCUCCAACAUAACAAAUUACGCGUACACAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUC ...-...((((((((.....(((........((((((....))))))....)))..........................((((((((....))))))))...)))))))) ( -25.20) >DroSec_CAF1 13680 107 + 1 CCU-UAAGGGUAAC---AUUGAUUAUCAAACAGAUGUCCUGACAUUUACAUAGCCCCAACAAAACAAAUUACGCUUAUUCAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUC ...-...(((((((---(.............((((((....)))))).................................((((((((....))))))))...)))))))) ( -23.60) >DroSim_CAF1 13297 101 + 1 CAU-UACA---------GUUGCUUAUCAAACAGAUGUCCUGUCAUUUACAUAGCCCCAACAAAACAAAUUACGCUUACUCAGGAUGGAGGUAUCCAUCCUUCCUGUUCCUC ...-.(((---------(..(((......((((.....)))).........)))..........................((((((((....))))))))..))))..... ( -18.06) >DroEre_CAF1 14861 107 + 1 CCG-UGAGGGUAAC---AUGGCUUGUCAAACAGAUGUCCCGGCAUUUACAUAGCCCCAAAAUAACAAAUUACGUUUGCUCAGGAUGGAGGUAUCCAUCCAUCCUGUUGCUC ((.-...)).....---..(((((((.....((((((....))))))))).)))).....................((.(((((((((((...)).)))))))))..)).. ( -32.00) >DroYak_CAF1 12863 108 + 1 CCUUUGGGGGUAAC---UUGUCUCAUCAAACAGAUGUCCUGGCAUUUACAUAGCCCCAAAAUAACAAAUUACGCUUACUCAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUC ..(((((((((((.---.((((.((((.....))))....)))).))))....)))))))............((..((..((((((((....))))))))....)).)).. ( -32.20) >DroAna_CAF1 14009 101 + 1 CGU-UAAAGGUGCUUCCUUCAAAUGCAAAGCAGAUGUUGAGUUGUUUACAUAACCCCAUCAUAGAAACUCAUUAUUAAACAGGAUGAAAGCAUCCU--------AUUUUU- ...-...(((((((((.(((...(((...)))((((.((((((......................)))))).)))).....))).).))))).)))--------......- ( -16.75) >consensus CCU_UAAGGGUAAC___AUUGCUUAUCAAACAGAUGUCCUGGCAUUUACAUAGCCCCAACAUAACAAAUUACGCUUACUCAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUC .......(((((((.................((((((....))))))..................................(((((((....))))))).....))))))) (-11.34 = -13.37 + 2.03)

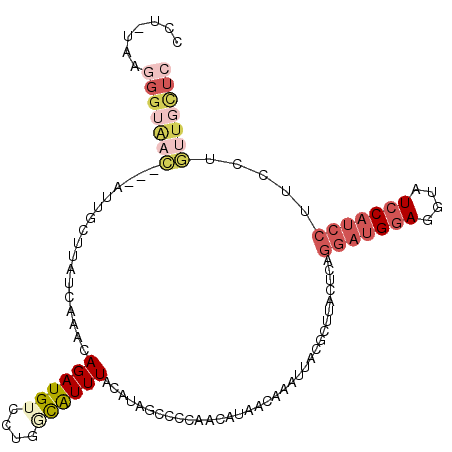
| Location | 5,594,407 – 5,594,518 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -24.62 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5594407 111 - 23771897 GUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGUGUACGCGUAAUUUGUUAUGUUGGAGCUAUGUAAAUGCC (((..((((((((((....)))))....((((((((....)))).(((...((((((((....)))))))).)))..((((((....)))))))))).)))))....))). ( -31.10) >DroSec_CAF1 13716 111 - 1 GUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGAAUAAGCGUAAUUUGUUUUGUUGGGGCUAUGUAAAUGUC .....((((((((((....))))).((((((.((((....)))).......((((((((....)))))))))))))).(.((((.......)))).).)))))........ ( -30.30) >DroSim_CAF1 13327 111 - 1 GUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUCGAGGAACAGGAAGGAUGGAUACCUCCAUCCUGAGUAAGCGUAAUUUGUUUUGUUGGGGCUAUGUAAAUGAC ....((((..(((((....))))).((((..((((..(....((((((((.((((((((....))))))))..(....)....))))))))..)..))))..)))))))). ( -31.40) >DroEre_CAF1 14897 111 - 1 GUGAUCAGGGAAUCCAAGAGGAUUAUUAUCCAGCUUUGUUGAGCAACAGGAUGGAUGGAUACCUCCAUCCUGAGCAAACGUAAUUUGUUAUUUUGGGGCUAUGUAAAUGCC .............((((((((((....)))).((((....))))..(((((((((.((...)))))))))))((((((.....)))))).))))))(((.........))) ( -33.30) >DroYak_CAF1 12900 111 - 1 GCAAUCAGGGAAUCCGAGAGGGUUGCUAUUCGGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGAGUAAGCGUAAUUUGUUAUUUUGGGGCUAUGUAAAUGCC .......((.(((((....))))).))....(((((((...(((..(((((((((((((....)))))))).....((((.....)))).)))))..)))...)))).))) ( -35.10) >consensus GUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGAGUAAGCGUAAUUUGUUAUGUUGGGGCUAUGUAAAUGCC .....((((((((((....)))))....((((((((((((....)))))).((((((((....))))))))....................)))))).)))))........ (-24.62 = -25.18 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:50 2006