
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,589,879 – 5,589,998 |
| Length | 119 |
| Max. P | 0.683534 |

| Location | 5,589,879 – 5,589,998 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5589879 119 - 23771897 AGCUGAAGUCCAAGUCAGCAGAGGAAGAAGAAGCAGAAGCAGUUAAAGCCAAUGACCGCAAGUAACUUACAUUUGAAAAAUGUGUGCAAAAAGCACAGCGAAAGGAGACUUGCAUACAG .(((((........)))))...((........((....))........)).......((((((..((((((((.....)))))((((.....)))).....)))...))))))...... ( -26.69) >DroSec_CAF1 9361 119 - 1 AGCUGAAGUCCAAGUCAGCAGAGGAAGAAGAAGCAGAAGCAGUUAAAGCCAAUGACCGCAAGUAACUUACAUUUGAAAAAUGUGUGCAAAAAGCACAGCGAAAGGAGACUUGCAUACAG .(((((........)))))...((........((....))........)).......((((((..((((((((.....)))))((((.....)))).....)))...))))))...... ( -26.69) >DroSim_CAF1 8780 119 - 1 AGCUGAAGUCCAAGUCAGCGGAGGAAGCAGAAGCAGAAGCAGUUAAAGCCAAUGACCGCAAGUAACUUACAUUUGAAAAAUGUGUGCAAAAAGCACAGCGAAAGGAGACUUGCAUACAG .(((((........)))))((.....((....))....((.......))......))((((((..((((((((.....)))))((((.....)))).....)))...))))))...... ( -27.60) >DroEre_CAF1 10426 112 - 1 AGCUGAAGUCCAAGUCAGCAGA-------GAAGAAGAAGCAGUUAAAGCCAAUGACCGCAAGUAACUUACAUUUGAAAAAUGUGUGCAAAAAGCACAGCGAAAGGAGACAUGCAUACAG .(((((........)))))...-------.........(((...............(((.........(((((.....)))))((((.....)))).)))...(....).)))...... ( -22.90) >DroYak_CAF1 8190 114 - 1 AGCUGAAGUCCAAGUCAGCAGA-----AUGAAGAAGAAGCAGUUAAAGGCAAUGACCGCAAGUAACUUACAUUUGAAAAAUGUGUGCAAAAAGCACAGCGAAAGGAGACUUGCAUACAG (((((...((....(((.....-----.)))....))..)))))...((......))((((((..((((((((.....)))))((((.....)))).....)))...))))))...... ( -25.80) >DroAna_CAF1 9637 102 - 1 AGAAGAAG-------CAGCA---------GCAGCAAAAGCAGUUAAAGCCAAUGACCGCAAGUAACUUACAUUUGAAAAAUGUGUGCAA-AAGCACAGCGAAAGGAGACUUGCAUACAG .......(-------(...(---------((.((....)).)))...))........((((((..((((((((.....)))))((((..-..)))).....)))...))))))...... ( -20.50) >consensus AGCUGAAGUCCAAGUCAGCAGA_____AAGAAGCAGAAGCAGUUAAAGCCAAUGACCGCAAGUAACUUACAUUUGAAAAAUGUGUGCAAAAAGCACAGCGAAAGGAGACUUGCAUACAG .(((((........)))))......................................((((((..((((((((.....)))))((((.....)))).....)))...))))))...... (-21.22 = -21.88 + 0.67)

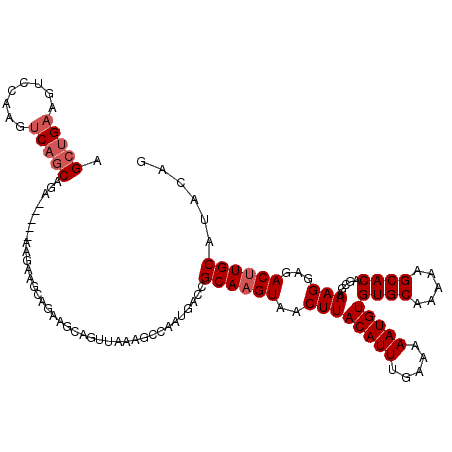
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:49 2006