| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
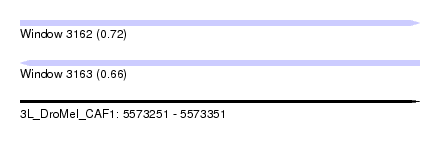
| Location | 5,573,251 – 5,573,351 |
| Length | 100 |
| Max. P | 0.717595 |

| Location | 5,573,251 – 5,573,351 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -22.41 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5573251 100 + 23771897 AAGGUGGAGGAGCUGGGCAUGGGAAACAAUAUCCAGGAGGACCAGGCGGUCCUGGUAAGCCUCUAGGCCCCUGGAUGAAAGGCAGAUUUGGGGGGUAAGG ....((((((..((((..((.(....).))..)))).((((((....))))))......))))))..((((..(((.........)))..))))...... ( -38.90) >DroSim_CAF1 6544 100 + 1 AAGGUGGAGCAGCCGGGCAUGGGAAACAAUAUCCAGGAGGACCAGGCGGUCCUGGUAAUCCUCUCGGUCCCUGGAUGAAAGGCAGAUUUGGGGGGUAAGG ..(((......)))((..((.(....).))..))((((..((((((....))))))..))))((.(.((((..(((.........)))..)))).).)). ( -34.40) >DroEre_CAF1 6189 100 + 1 AUGGUGGAGCAGCCGGGCAUGGGAAACAAUAUCCAGGAGGACCCGGCGGUCCUGGUAGGCCUCUAGGUCCCUGAAGGAAAGGCAGAUUUGGGGGGUAAGG .....(((.(.((((((....(....)....(((....)))))))))).))).......(((((((((((((.......)))..))))))))))...... ( -36.40) >DroWil_CAF1 7824 100 + 1 AUGGUGGUGCUGCCGGGCAGGGGAAACAGUAUCCUGGUGGACCCGGUGGUCCUGGCAUGCCUCGCGGUCCGGGUAUUAUUUGUACAUUCUGAUUGAAUGG ..(((((((((((((((....(....).....))))))((((((((.(((........)))))).))))).)))))))))....(((((.....))))). ( -36.70) >DroYak_CAF1 6536 100 + 1 AUGGCGGACCAGCCGGGCAGGGGAAACAAUAUCCAGGAGGACCCGGUGGUCCUGGUAAGCCUCUGGGUCCCUGAAUGAAUGGCAGAUUUGGGGGAUAAGG ..(((((((((.(((((....(....)....(((....))))))))))))))......))).((...((((..(((.........)))..))))...)). ( -37.40) >DroAna_CAF1 6478 100 + 1 CUGGCGGAAUGGCUGGGCAGGGGAAGCAAUAUCCAGGAGGUCCCGGUGGACCAGGUAGACCUCUAGGCCCCGGGAUAAUCGGAACAUUCGGUGGAAAAGG .(.((.(((((.((((..(..(....)..)..))))((.(((((((.((.(((((....)))...)))))))))))..))....))))).)).)...... ( -36.20) >consensus AUGGUGGAGCAGCCGGGCAGGGGAAACAAUAUCCAGGAGGACCCGGCGGUCCUGGUAAGCCUCUAGGUCCCUGGAUGAAAGGCAGAUUUGGGGGGUAAGG ..((.((....(((((..(..(....)..)..))....(((((....))))).)))...)).))...(((((((((.........)))))))))...... (-22.41 = -22.50 + 0.09)



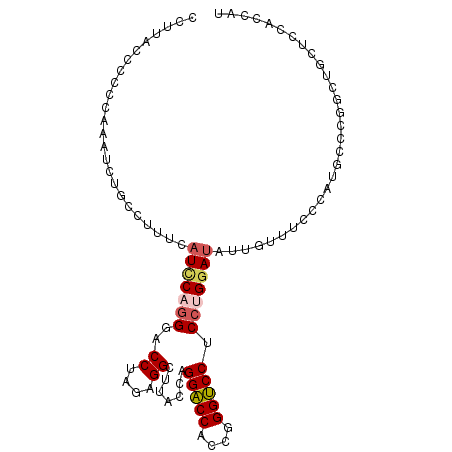
| Location | 5,573,251 – 5,573,351 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.97 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5573251 100 - 23771897 CCUUACCCCCCAAAUCUGCCUUUCAUCCAGGGGCCUAGAGGCUUACCAGGACCGCCUGGUCCUCCUGGAUAUUGUUUCCCAUGCCCAGCUCCUCCACCUU ........................(((((((((((....))))....((((((....))))))))))))).............................. ( -25.20) >DroSim_CAF1 6544 100 - 1 CCUUACCCCCCAAAUCUGCCUUUCAUCCAGGGACCGAGAGGAUUACCAGGACCGCCUGGUCCUCCUGGAUAUUGUUUCCCAUGCCCGGCUGCUCCACCUU .................(((....((((((((.((....))...((((((....))))))..))))))))...((.......))..)))........... ( -27.10) >DroEre_CAF1 6189 100 - 1 CCUUACCCCCCAAAUCUGCCUUUCCUUCAGGGACCUAGAGGCCUACCAGGACCGCCGGGUCCUCCUGGAUAUUGUUUCCCAUGCCCGGCUGCUCCACCAU .................((((((((.....)))....)))))......((((.(((((((......(((.......)))...))))))).).)))..... ( -28.30) >DroWil_CAF1 7824 100 - 1 CCAUUCAAUCAGAAUGUACAAAUAAUACCCGGACCGCGAGGCAUGCCAGGACCACCGGGUCCACCAGGAUACUGUUUCCCCUGCCCGGCAGCACCACCAU .(((((.....)))))..............((..(....)((.((((.(((((....)))))..((((...........))))...)))))).))..... ( -27.90) >DroYak_CAF1 6536 100 - 1 CCUUAUCCCCCAAAUCUGCCAUUCAUUCAGGGACCCAGAGGCUUACCAGGACCACCGGGUCCUCCUGGAUAUUGUUUCCCCUGCCCGGCUGGUCCGCCAU .....((((....................))))......(((......((((((((((((......(((.......)))...)))))).))))))))).. ( -30.05) >DroAna_CAF1 6478 100 - 1 CCUUUUCCACCGAAUGUUCCGAUUAUCCCGGGGCCUAGAGGUCUACCUGGUCCACCGGGACCUCCUGGAUAUUGCUUCCCCUGCCCAGCCAUUCCGCCAG ...........(((((...((.......))((((...(((((...(((((....))))))))))..(((.......)))...))))...)))))...... ( -26.80) >consensus CCUUACCCCCCAAAUCUGCCUUUCAUCCAGGGACCUAGAGGCUUACCAGGACCACCGGGUCCUCCUGGAUAUUGUUUCCCAUGCCCGGCUGCUCCACCAU ........................(((((((..((....)).......(((((....))))).))))))).............................. (-16.58 = -16.97 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:40 2006