
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
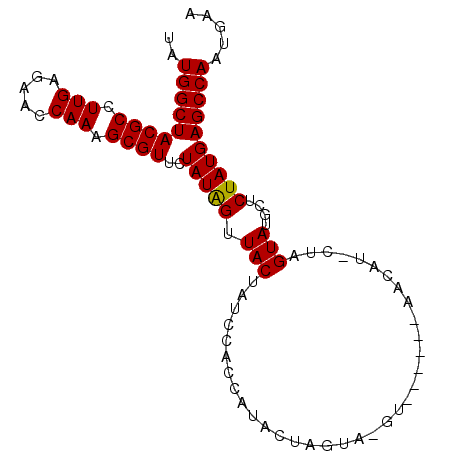
| Location | 5,521,328 – 5,521,423 |
| Length | 95 |
| Max. P | 0.898687 |

| Location | 5,521,328 – 5,521,423 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -13.29 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5521328 95 + 23771897 UAUGGCUACGCCUUGAGAACCAAAGCGUUCUAUAGUUACUAUCCAAUAUACUCGUA-GAUGCACUGAAGUGGUAGUAUGCUCUAUGAGCCAAUGAA (((((..((((.(((.....))).)))).)))))................((((((-((.((((((......))))..))))))))))........ ( -22.10) >DroSec_CAF1 82996 89 + 1 UAUGGCUACGCCUUGAGAACCAAAGCGUUCUAUAGUUACUAUGCACCAUACUAGUA-GU------AACAUACUUGUAUAUACUAUGAGCCAAUGAA ..(((((((((.(((.....))).))))..((((((...((((((.......((((-..------....)))))))))).)))))))))))..... ( -21.51) >DroSim_CAF1 84739 89 + 1 UAUGGCUACGCCUUGAGAACCAAAGCGUUCUAUAGUUACUAUGCACCAUACUAGUA-GU------AACAUACUUGUAUAUACUAUGAGCCAAUGAA ..(((((((((.(((.....))).))))..((((((...((((((.......((((-..------....)))))))))).)))))))))))..... ( -21.51) >DroEre_CAF1 83647 75 + 1 CAUGGCUACGCCUUGAGAACCAAAGCGUUCUAUGGGUACCAUCCAGCAUGC---------------------UAGUAUGCUCUAUGAGCCAAUGAA ..(((((..((((..(((((......)))))..))))..(((..((((((.---------------------...))))))..))))))))..... ( -24.00) >DroYak_CAF1 71912 88 + 1 UAUGGCUACGCCUUGAGAACCAAAGCGUUCUAUGGUUACUAUCCAGUAUGU-AGUAUGU------AGUAU-GUAGUAUGCUCUAUGAGCCAAUGAA ..(((((((((.(((.....))).))))..(((((.((((((..(.(((((-...))))------).)..-))))))....))))))))))..... ( -19.90) >consensus UAUGGCUACGCCUUGAGAACCAAAGCGUUCUAUAGUUACUAUCCACCAUACUAGUA_GU______AACAU_CUAGUAUGCUCUAUGAGCCAAUGAA ..(((((((((.(((.....))).))))..(((((.(((...................................)))....))))))))))..... (-13.29 = -13.05 + -0.24)

| Location | 5,521,328 – 5,521,423 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.28 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5521328 95 - 23771897 UUCAUUGGCUCAUAGAGCAUACUACCACUUCAGUGCAUC-UACGAGUAUAUUGGAUAGUAACUAUAGAACGCUUUGGUUCUCAAGGCGUAGCCAUA .....((((((((((....((((((((.....((((...-.....))))..))).))))).)))).))((((((((.....)))))))).)))).. ( -23.70) >DroSec_CAF1 82996 89 - 1 UUCAUUGGCUCAUAGUAUAUACAAGUAUGUU------AC-UACUAGUAUGGUGCAUAGUAACUAUAGAACGCUUUGGUUCUCAAGGCGUAGCCAUA .....(((((....((((((((.((((....------..-)))).)))).))))..............((((((((.....))))))))))))).. ( -24.20) >DroSim_CAF1 84739 89 - 1 UUCAUUGGCUCAUAGUAUAUACAAGUAUGUU------AC-UACUAGUAUGGUGCAUAGUAACUAUAGAACGCUUUGGUUCUCAAGGCGUAGCCAUA .....(((((....((((((((.((((....------..-)))).)))).))))..............((((((((.....))))))))))))).. ( -24.20) >DroEre_CAF1 83647 75 - 1 UUCAUUGGCUCAUAGAGCAUACUA---------------------GCAUGCUGGAUGGUACCCAUAGAACGCUUUGGUUCUCAAGGCGUAGCCAUG .....((((((((..(((((....---------------------..)))))..)))...........((((((((.....))))))))))))).. ( -23.30) >DroYak_CAF1 71912 88 - 1 UUCAUUGGCUCAUAGAGCAUACUAC-AUACU------ACAUACU-ACAUACUGGAUAGUAACCAUAGAACGCUUUGGUUCUCAAGGCGUAGCCAUA .....(((((...............-.....------...((((-((......).)))))........((((((((.....))))))))))))).. ( -18.10) >consensus UUCAUUGGCUCAUAGAGCAUACUAC_AUGUU______AC_UACUAGUAUGCUGGAUAGUAACUAUAGAACGCUUUGGUUCUCAAGGCGUAGCCAUA .....(((((............................................((((...))))...((((((((.....))))))))))))).. (-15.32 = -15.28 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:25 2006