
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,518,950 – 5,519,058 |
| Length | 108 |
| Max. P | 0.898642 |

| Location | 5,518,950 – 5,519,058 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
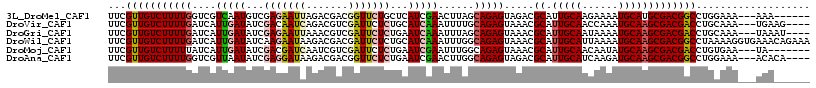
>3L_DroMel_CAF1 5518950 108 + 23771897 UUCGUUGUCUUUUGGUCGUCAAUGUCGAGAAUUAGACGACGGUUCUGCUCAUCGAACUUAGCAGAGUAGACGCAUUGCAAGAAAAUGCAUGCGACGGCCUGGAAA---AAA------ .......(((...(((((((..(((((.........)))))..(((((((.............))))))).((((.(((......)))))))))))))).)))..---...------ ( -32.72) >DroVir_CAF1 64640 110 + 1 UUCGUUGUCUUUUGAUCAUUGAUAUCGACAAUCAGACGUCGAUUCUCUGCAUCAAAUUUUGCAGAGUAAACGCAUUGCAACCAAAUGCAAGCGACGACCUGCAAA---UGAAG---- ((((((....((((((((..((.((((((........))))))))..)).))))))...(((((.((...(((.(((((......))))))))...)))))))))---)))).---- ( -34.40) >DroGri_CAF1 40827 110 + 1 UUCGUUGUCUUUUGAUCAUUGAUAUCGAGAAUUAAACGUCGAUUCUCUGAAUCAAAUUUAGCAGAGUAAACGCAUUGCAAUAAAAUGCAAGCGACGACCUGCAAA---UAAAU---- ..................(((((.(((((((((.......))))))).))))))).....((((.((...(((.(((((......))))))))...))))))...---.....---- ( -26.90) >DroWil_CAF1 60592 117 + 1 UUCGUUGUCUUUUGAUCAUUGAUAUCAAGAAUAAGACGACGAUUCUCUGCAUCAAAUUUGGCAGAGUAAACGCAUUGCAUUAAAAUGCAAGCGACGGCCUAAAAGGUGAAACAGAAA .((((((((((....((.(((....)))))..))))))))))...((((..(((..((((((...((...(((.((((((....))))))))))).)))..)))..)))..)))).. ( -35.00) >DroMoj_CAF1 77010 107 + 1 UUCGUUGUCUUUUUAUCAUUGAUAUCGACGAUCAAUCGUCGAUUCUCUGAAUCGAAUUUGGCAGAGUAAACGCAUUGCAACAAUAUGCAAGCGACGACCUGUGAA---UA------- ...............(((..((.((((((((....))))))))))..))).......((.((((.((...(((.(((((......))))))))...)))))).))---..------- ( -29.60) >DroAna_CAF1 42934 110 + 1 UUCGUUGUCUUUUGGUCGUUAAUAUCGAGGAUAAGACGACGGUUCUCUGAAUCGAACUUGGCAGAGUAGACGCAUUGCAUCAAGAUGCAAGCGACGGCCUGGAAA---ACACA---- (((((((((((.(..(((.......)))..).))))))))(((((...))))))))((.(((...((...(((.((((((....))))))))))).))).))...---.....---- ( -31.80) >consensus UUCGUUGUCUUUUGAUCAUUGAUAUCGAGAAUAAGACGACGAUUCUCUGAAUCAAAUUUGGCAGAGUAAACGCAUUGCAACAAAAUGCAAGCGACGACCUGCAAA___AAA______ ...(((((((((((....(((((...(((((((.......)))))))...)))))......))))).....((.(((((......)))))))))))))................... (-19.14 = -19.62 + 0.48)


| Location | 5,518,950 – 5,519,058 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -14.35 |
| Energy contribution | -14.30 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
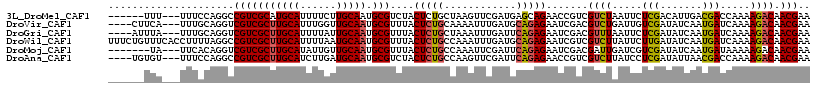
>3L_DroMel_CAF1 5518950 108 - 23771897 ------UUU---UUUCCAGGCCGUCGCAUGCAUUUUCUUGCAAUGCGUCUACUCUGCUAAGUUCGAUGAGCAGAACCGUCGUCUAAUUCUCGACAUUGACGACCAAAAGACAACGAA ------...---.........(((.(((((((......))).))))((((..((((((..........))))))...((((((..............))))))....)))).))).. ( -28.44) >DroVir_CAF1 64640 110 - 1 ----CUUCA---UUUGCAGGUCGUCGCUUGCAUUUGGUUGCAAUGCGUUUACUCUGCAAAAUUUGAUGCAGAGAAUCGACGUCUGAUUGUCGAUAUCAAUGAUCAAAAGACAACGAA ----..((.---(((((((((...((((((((......))))).)))...)).))))))).((((((.((..(((((((((......))))))).))..)))))))).......)). ( -35.40) >DroGri_CAF1 40827 110 - 1 ----AUUUA---UUUGCAGGUCGUCGCUUGCAUUUUAUUGCAAUGCGUUUACUCUGCUAAAUUUGAUUCAGAGAAUCGACGUUUAAUUCUCGAUAUCAAUGAUCAAAAGACAACGAA ----.....---...((((((...((((((((......))))).)))...)).))))....((((((.((..(((((((.(......).))))).))..)))))))).......... ( -26.50) >DroWil_CAF1 60592 117 - 1 UUUCUGUUUCACCUUUUAGGCCGUCGCUUGCAUUUUAAUGCAAUGCGUUUACUCUGCCAAAUUUGAUGCAGAGAAUCGUCGUCUUAUUCUUGAUAUCAAUGAUCAAAAGACAACGAA (((((((.(((...(((.(((.(.(((((((((....)))))).)))......).))))))..))).))))))).((((.(((((....(((((.......)))))))))).)))). ( -33.30) >DroMoj_CAF1 77010 107 - 1 -------UA---UUCACAGGUCGUCGCUUGCAUAUUGUUGCAAUGCGUUUACUCUGCCAAAUUCGAUUCAGAGAAUCGACGAUUGAUCGUCGAUAUCAAUGAUAAAAAGACAACGAA -------..---........((((.(((((((......))))).))((((.(((((............))))).((((((((....)))))))).............)))).)))). ( -27.30) >DroAna_CAF1 42934 110 - 1 ----UGUGU---UUUCCAGGCCGUCGCUUGCAUCUUGAUGCAAUGCGUCUACUCUGCCAAGUUCGAUUCAGAGAACCGUCGUCUUAUCCUCGAUAUUAACGACCAAAAGACAACGAA ----(((.(---(((...((..(.(((((((((....)))))).))).)..(((((............)))))..))(((((..((((...))))...))))).)))).)))..... ( -26.50) >consensus ______UUA___UUUCCAGGCCGUCGCUUGCAUUUUGUUGCAAUGCGUUUACUCUGCCAAAUUCGAUGCAGAGAAUCGACGUCUAAUUCUCGAUAUCAAUGAUCAAAAGACAACGAA .....................(((((((((((......))))).)))....(((((............))))).......((((.....(((.......))).....)))).))).. (-14.35 = -14.30 + -0.05)
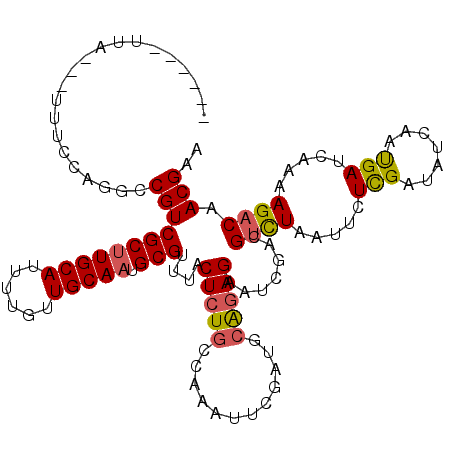
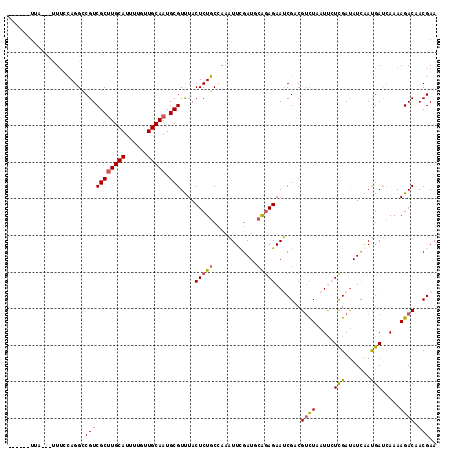
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:23 2006